PanelApp and the NHS Genomic Medicine Service (GMS) Panels Resource
Here on the PanelApp website you can find all the gene panels that relate to genomic tests listed in the NHS National Genomic Test Directory, as well as the virtual gene panels that were used in the 100,000 Genomes Project. Not all the genes and panels shown here are available to be chosen for testing as part of the NHS Genomic Medicine Service (GMS).

|
NHS users should view the set of genes/panels that are available for testing as part of the GMS on the
NHS GMS Panels Resource
website. These are the panels that can be chosen when requesting a genomic test. The green (diagnostic evidence level) genes shown on these panels are those that are analysed as part of the diagnostic pathway*. The panel versions are periodically updated from PanelApp.
|
* please note that some genes, in particular non-coding, mitochondrial and those located in difficult to sequence genomic regions, may not be accurately analysed via the GMS WGS service for rare and inherited diseases due to limitations of the WGS analysis pipeline or WGS technology. These tests are outside the scope of Genomics England's UKAS ISO 15189 accreditation. For full details on GMS WGS limitations, please refer to the Rare Disease Analysis User Guide available in NHS Futures.
On the NHS GMS Panels Resource website you can:
- Browse the approved/signed-off versions of panels in use in the NHS GMS
- Find which panel(s) a gene is on
- Find the content of a panel associated with a clinical indication
On the PanelApp website you can:
- Browse all panels
- Leave a review on any panel to suggest a change of rating, mode of inheritance or a new gene that should be added to the panel
- See the latest versions of all panels (Note panel versions used in the GMS may differ from the current versions listed on PanelApp, therefore the NHS GMS Panels Resource should be used to query panel content in current use within the GMS)
See the PanelApp Handbook V105
for more information.
Known issues: Please visit the 'Known issues in PanelApp functionality and recommended workarounds' section of the Contact, Content & Glossary page for more information.
29th July 2021: New PanelApp publication
Read our new publication in the American Journal of Human Genetics highlighting the benefits of using the PanelApp platform in harmonising gene and panel curation efforts across healthcare systems in collaboration with PanelApp Australia.
Scaling national and international improvement in virtual gene panel curation using a collaborative approach to discordance resolution, AJHG, 2021
May 2021: NHS GMS Panels Resource Now Available!

We are pleased to announce the launch of the NHS Genomic Medicine Service (GMS) Panels Resource
, which offers a view of the 173 signed-off panels that relate to genomic tests listed in the NHS National Genomic Test Directory. This platform contains only ‘Green’ (diagnostic level of evidence) genes, STRs, and regions (CNVs) that have been approved for diagnostic testing in the NHS in England and is intended for use by NHS users and clinicians.
The main Genomics England PanelApp knowledgebase here will be dedicated for use by the wider scientific community for visualisation and reviewing of genes/genomic entities on virtual gene panels and will continue to be updated by Genomics England curators.
For more information on the evaluation and approval process of the content of GMS signed-off panels, please refer to the updating the NHS National Genomic Test Directory webpage.
Click here to go to the NHS GMS Panels Resource
What is PanelApp?
Genomics England PanelApp is a publicly-available knowledgebase that allows virtual gene panels related to human disorders to be created, stored and queried. It includes a crowdsourcing tool that allows genes and genomic entities (short tandem repeats/STRs and copy number variants/CNVs) to be added or reviewed by experts throughout the worldwide scientific community, providing an opportunity for the standardisation of gene panels, and a consensus on which genes have sufficient evidence for disease association.
Diagnostic-grade ‘Green’ genes/genomic entities, and their modes of inheritance are used in genome interpretation. Originally developed to aid interpretation of participant genomes in the 100,000 Genomes Project, PanelApp is now also being used as the platform for achieving consensus on gene panels in the NHS Genomic Medicine Service (GMS). As panels in PanelApp are publicly available, they can also be used by other groups and projects.
Find out more about PanelApp
Read the PanelApp publication in Nature Genetics: PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels
Read the PanelApp Handbook V105
Read the PanelApp Reviewer's Guide
Watch PanelApp Videos
Register to be a reviewer
Query The new PanelApp API is available here
PanelApp Accreditation

PanelApp is accredited under the Genomics England ISO 15189 Accreditation Schedule. Please refer to the Schedule for further details.
Types of PanelApp Gene Panels
A ‘Panel Type’ is assigned to each PanelApp panel that denotes what the gene panel is utilised for. Panels may have more than one panel type, for example when the panel was created for both the 100,000 Genomes Project and the NHS Genomic Medicine Service. Current panel types are:
Rare Disease 100K: a gene panel used for the Rare Disease programme of the 100,000 Genomes Project.
Cancer Germline 100K: a gene panel used for the Germline Cancer programme of the 100,000 Genomes Project.
GMS Rare Disease: a panel developed for the NHS Genomic Medicine Service; may be delivered by whole exome sequencing, whole genome sequencing, or as a ‘wet lab’ panel.
GMS Rare Disease Virtual: a panel developed for the NHS Genomic Medicine Service; will be used as a virtual panel for whole genome sequencing.
GMS Cancer Germline Virtual: a panel developed for the NHS Genomic Medicine Service for analysis of germline cancer susceptibility from whole genome sequencing (WGS).
Super Panel: A panel made up of two or more component panels. When the component panels are updated, the Super panel is automatically updated.
Component of Super Panel: A panel that is a constituent of a Super panel. Changes to this panel will automatically update the Super panel.
Actionable: A panel containing actionable information related to genes or genomic entities, such as clinical trial information.
Research: A panel from a research project.
External Diagnostic Lab: a gene panel from a diagnostic lab or other source, external to Genomics England and not directly used for genome analysis for the 100,000 Genomes Project or NHS Genomic Medicine Service.
How Gene Panels were Defined and Created for the 100,000 Genomes Project
For the 100,000 Genomes Project, gene panels are mapped to one or more recruitment categories, indicated by the gene panel name (Level 4 Title) and/or listed under ‘relevant disorders’ for each panel in PanelApp.
For the 100,000 Genomes Rare Disease programme, gene panels are created in the following steps:
-
An initial gene list is drawn up from established sources (UKGTN, Radboud UMC, Emory Genetic Laboratory and Illumina) and from disease area experts. The initial gene panel created is Version 0.
-
Expert review of each gene is crowdsourced.
-
Evaluation of the reviews, further curation and consultation with the Genomics England clinical team results in a finalised panel.
-
Promotion of the panel to Version 1 allows the panel to be used in the interpretation of participant genomes.
For the 100,000 Genomes Cancer Germline programme, gene panels are created in the following steps:
-
Initial gene lists with strong clinical evidence conferring susceptibility of clinically-relevant penetrance to the respective tumour type were submitted from the gene lists used in the interpretation pipeline. Initial gene panels are Version 0.
-
Review, Evaluation and Promotion of the panel as outlined in steps 2-4 for the Rare Disease programme, above.
Note that PanelApp gene panels are not used in interpretation until initial expert review and curation has been completed and the panel has been promoted to Version 1. Before this, panels are viewable and can be reviewed but the rating of genes has not been finalised.
How Gene Panels are Defined for the NHSE Genomic Medicine Service
Panels for the Genomic Medicine Service (GMS) are mapped to Clinical Indications specified in the National Genomic Test Directory. The Clinical Indication (E.g. R59 Early onset or syndromic epilepsy) is listed in the Description box, and the Clinical Indication code (R59) is listed in the ‘Relevant disorder’ field. Consensus gene/entity lists are reached in consultation with experts from Genomic Laboratory Hubs (GLHs) around the country.
Entities on a Gene Panel
A PanelApp panel can contain the following genomic entities:
-
Gene
-
STR (Short Tandem Repeat). STRs can be disease-causing when a particular number of repeats is present.
-
CNV (Copy Number Variant) from the curated ClinGen Dosage Sensitivity Map curated regions. PanelApp currently supports region-loss and region-gain.
Green genomic entities on a version 1+ panel will be used for genome interpretation in the 100,000 Genomes Project and NHSE Genomic Medicine Service. For further details on the information captured for genes, STRs and CNVs, see the PanelApp Handbook V105.
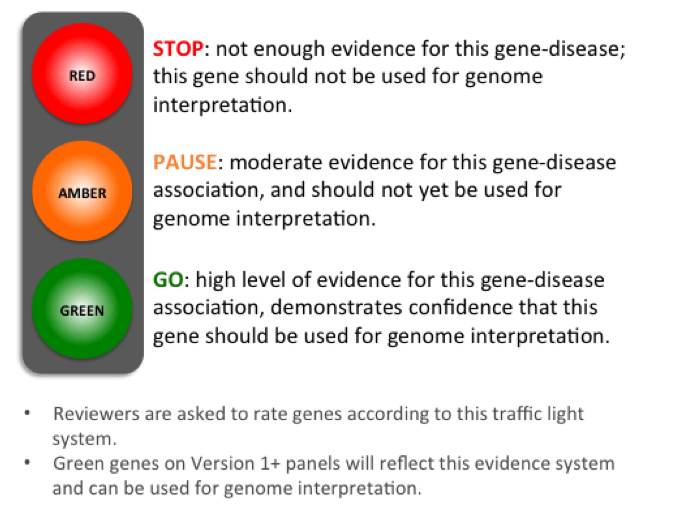
Understanding Gene Ratings on a Version 1+ Gene Panel
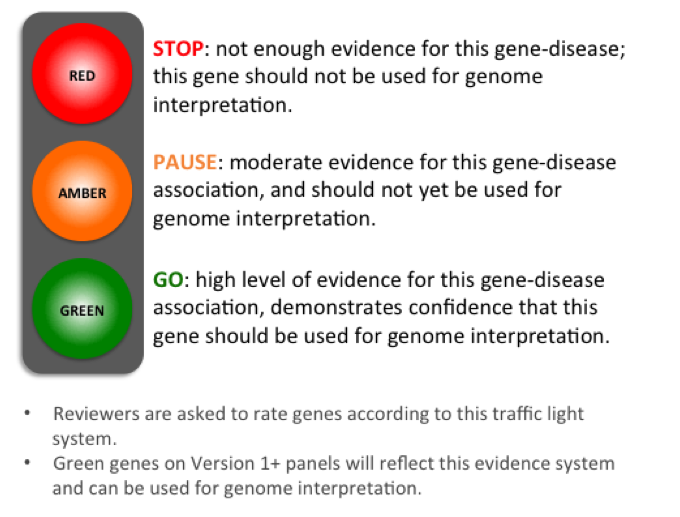
Figure 1: A traffic-light system is used to rate genes on a Version 1+ gene panel.
We classify genes on a panel according to a traffic light system (Figure 1). Genes are rated in terms of the level of evidence to support their association with the phenotypes covered by the gene panel in question. For Rare Disease, the criteria for assessing the evidence were developed from a combination of the ClinGen DEFINITIVE and DDG2P CONFIRMED gene evidence levels and can be viewed on the Guidelines tab.
A diagnostic-grade (Green) rating on a Version 1+ panel requires evidence from 3 or more unrelated families or from 2-3 unrelated families where there is strong additional functional data. Genes that do not meet these criteria are rated as Amber (borderline) or Red (low level of evidence) and are not used in Tiering.
For Cancer Germline panels, the Green genes on a Version 1+ panel are those with strong clinical evidence conferring susceptibility of clinically-relevant penetrance to the respective tumour type.
How Do I Add Genes, Genomic Entities and Reviews to a Gene Panel?
PanelApp has more than 250 active reviewers from over 25 countries. We encourage experts to contribute their knowledge to update existing panels and help create new diagnostic-grade panels - please register. Please refer to the PanelApp Handbook V105 for a step-by-step guide to reviewing gene panels.
Uses and Users of PanelApp
For the 100,000 Genomes Project Rare Disease Programme and/or the NHS Genomic Medicine Service, the diagnostic grade ‘Green’ genes/genomic entities, and their modes of inheritance on the Version 1+ PanelApp virtual gene panels are used for genome interpretation. For Rare Diseases, the tiering process involving virtual gene panels aids evaluation of findings by annotating variants that are plausibly pathogenic based on their segregation in the family, frequency in control populations, and effect on protein coding. Variants in diagnostic-grade ‘Green’ genes can be tiered as Tier 1 or Tier 2.
In the Genomics England 100,000 Genomes Project Cancer Programme, germline variants are tiered as Tier 1 if pathogenic or likely pathogenic variants are found in Green genes on the relevant cancer panel assigned based on the patient’s tumour type. Tier 3 variants are rare variants reported in Green genes on a broader set of cancer panels. For Tier 1 and Tier 3 variants, clinical review and confirmation of pathogenicity are essential and should be undertaken locally. This process is explained in more detailed in the technical information available here
Gene panels in PanelApp can also be utilised for other projects, clinics and databases:
I am a Clinician or other Healthcare Professional…
You could use PanelApp to:
• View and interpret a panel that has been applied to your patient
• Look at the evidence for inclusion of a gene(s) in your patient report during a MDT
• Review gene(s) on panel(s) for disease(s) matching your expertise
• Add diagnostic genes missing from a panel
• Add your publications as evidence for gene-disease relationships
I am a Researcher…
You could use PanelApp to:
• Source data for hypothesis generation.
• Study gene-disease relationships for genome interpretation, pathway analysis and more.
• Use ‘tagged’ genes to investigate genes with interesting disease-causing mechanisms.
• Use Red and Amber genes as a source of genes needing further evidence/investigation within your disease of interest for certain diseases.
• Review genes and/or panels you have expertise in to help genome interpretation.
• Add novel genes to panels that you find within your research.
• Add your publications as evidence for gene-disease relationships.
• Query PanelApp data through API.
I am a Bioinformatician
You could use PanelApp to:
• Use panels for your exome/genome interpretation pipeline.
• Query PanelApp data through API.
This is interim information and has not yet received final approval from Genomics England internal governance processes or NHS England and is therefore potentially subject to change.
Disclaimer
PanelApp Uses Exclusions of Liability
PanelApp gene lists are provided by Genomics England in good faith, and for the benefit of the research community. The original gene lists have been supplied through commercial and academic providers, but have not been separately verified by Genomics England. Equally, expert reviewers and curators adding content and comments through PanelApp do so under their own responsibility and without verification by Genomics England. Users must themselves verify the accuracy and content any information (including in respect of ownership of any intellectual property rights) obtained through PanelApp in advance of its use for any purpose. Genomics England, any expert reviewers and curators hereby exclude any and all liability, including without limitation under any laws of contract, tort (including negligence) or statutory duty or otherwise, and do not accept any liability or responsibility for uses made of the PanelApp gene lists or comments by individual reviewers.
PanelApp Connections Exclusions of Liability
Access to the PanelApp is not guaranteed. Genomics England accepts no liability and excludes all liability in respect of interruptions in accessing or inability to access the PanelApp gene lists at any time. Persons accessing PanelApp are responsible keep their anti-virus and security software up to date. Genomics England consequently accepts no liability for any loss or damage occurring as a result of any person using or connecting to or through PanelApp.
General Exclusion of Liability and Limitations on exclusions of Liability
Genomics England shall not be responsible for any of the following losses to persons using or accessing PanelApp, howsoever incurred, whether in contract, tort (including negligence), breach of statutory duty or otherwise: indirect losses, consequential losses, loss of income or revenue, loss of profit, third party claims, loss of business, loss of data, loss of anticipated savings, or any loss of opportunity.
No provision of this disclaimer shall operate to limit or exclude liabilities which cannot by the applicable laws of England be so limited or excluded.
PanelApp News
22nd October 2025: Updates to PanelApp web pages
We have refreshed the PanelApp web pages to bring users the most up-to-date information.
The latest changes include:
30th April 2025: Updates to GMS panels
We are pleased to announce our sixth major update to the NHS Genomic Medicine Service (GMS) panels. These updates align with NHS National Genomic Test Directory v8.
We have signed-off 113 NHS GMS panels in this update including 46 WGS panels and 67 non-WGS panels. This update consists of around 550 data changes to green entities across 84 GMS panels including significant updates to the R21, R412 Fetal anomalies and DDG2P panels. The remaining 29 panels are signed-off due to panel-level changes (see below), because they are components of superpanels in which other components have changes, or are superpanels.
In addition to data changes to green entities, panel names have been updated for three non-WGS panels in PanelApp.
- R93 from ‘Thalassaemia and other haemoglobinopathies’ to ‘Sickle cell, thalassaemia and other haemoglobinopathies’
- R213 from ‘PTEN Hamartoma Tumor Syndrome’ to ‘PTEN Hamartoma Tumour Syndrome’
- R361 from ‘Haemoglobinopathy trait or carrier testing’ to ‘Sickle cell, thalassaemia and other haemoglobinopathies trait or carrier testing’
This release would not have been possible without the efforts of our NHS partners and community of expert reviewers.
30th October 2024: Updates to GMS panels
We have now made our fifth major update to the NHS Genomic Medicine Service (GMS) panels. This is the third data update in the last six months.
We have signed-off 69 NHS GMS panels in this update. This includes over 400 data changes to green entities across 31 GMS panels including R21, R412 Fetal anomalies and DDG2P panels. The remaining 38 panels are signed-off due to panel-level changes (see below), because they are components of superpanels in which other components have changes, or are superpanels.
In addition to data changes to green entities, following panels have been newly added in PanelApp.
- R444 NICE approved PARP inhibitor treatment
- R446 APOL1 kidney donor testing
The other panel level changes include:
- R257 Unexplained young onset end-stage renal disease panel changed to a superpanel with 10 other renal panels and a newly added panel named ‘Unexplained young onset end-stage renal disease - additional genes’ as components. This is to simplify maintenance of this panel as an all-encompassing renal panel.
- The ‘additional genes’ panel contains entities previously present in R257 and not present in other component panels.
- Relevant disorder ‘Arthrogrythsis’ removed from R83 Arthrogryposis.
We would like to thank our NHS partners and our community of expert reviewers for their immense contributions to this data update.
7th August 2024: Updates to GMS panels
We are pleased to announce our fourth major update to the NHS Genomic Medicine Service (GMS) panels. These updates align with NHS National Genomic Test Directory v7.
We have signed-off 74 NHS GMS panels including nine super panels in this update. This update includes over 500 data changes to green entities across 44 GMS panels. The remaining 30 panels are signed-off due to panel-level changes (see below), because they are components of superpanels in which other components have changes, or are superpanels.
In addition to data changes to green entities, following panels have been added in PanelApp:
- R453 Monogenic short stature, formed from the gene content of R147 Growth failure in early childhood. The panel for R147 has been retired.
- R449 Diagnostic testing for Glutaric acidaemia I
- R450 Diagnostic testing for Isovaleric acidaemia
- R451 Diagnostic testing for MCADD - Medium-chain acyl-CoA dehydrogenase deficiency – full ACADM sequencing
The other panel level changes include:
- R195 Proteinuric renal disease has been made ready for the move to whole-genome sequencing (WGS)
- Panel name changes:
- R358 from 'Rhabdoid tumour predisposition' to 'Familial rhabdoid tumours'
- R98 from 'Likely inborn error of metabolism - targeted testing not possible' to 'Likely inborn error of metabolism'
- Screening panels including R275 Glutaric acidaemia I and R403 MCADD - Medium-chain acyl-CoA dehydrogenase deficiency - full ACADM sequencing have been retired.
This release would not have been possible without the efforts of our NHS partners and community of expert reviewers.
1st May 2024: Updates to GMS panels
We have now made the third major update to the NHS Genomic Medicine Service (GMS) panels. These updates align with NHS National Genomic Test Directory v6.
We have signed-off 69 NHS GMS panels including 10 superpanels in this update. This includes 1,155 data changes to green entities across 45 GMS panels including R21, R412 Fetal anomalies and DDG2P panels. The remaining 24 panels are signed-off due to panel-level changes (see below), changes to Ensembl IDs to align with Ensembl release 107 or because they are components of superpanels in which other components have changes.
The DDG2P panel is obtained from Gene2Phenotype (G2P) database and forms part of R27 Paediatric disorders superpanel. This update included re-mapping of terminologies between PanelApp and G2P as G2P terminology now aligns with nomenclature from Gene Curation Coalition (GenCC). This update added 250 additional green genes to the Paediatric disorders superpanel, 95 of which are not green on any other GMS panels.
In addition to data changes to green entities, panel level changes were made for the following panels:
- Panel name changed for R29 from ‘Intellectual disability – microarray and sequencing’ to ‘Intellectual disability’ and for R146 from ‘Disorders of sex development’ to ‘Differences of sex development’
- New component panels added to R27 Paediatric disorders and R441 Unexplained death in infancy and sudden unexplained death in childhood superpanels
- Ensembl IDs updated in PanelApp API for four genes (CCDC39, LRTOMT, SCO2, TBCE) that are green on GMS panels to align with Ensembl genome version 107
We would like to thank our NHS partners and our community of expert reviewers for their immense contributions to this data update.
20th December 2023: Updates to small non-WGS panels
Today, we have made minor updates to small non-Whole Genome Sequencing (WGS) panels for the NHS Genomic Medicine Service (GMS) to be aligned with NHS National Genomic Test Directory v5. This has been achieved with our partners at NHS England.
This update includes the following changes:
- R439 Fuchs endothelial corneal dystrophy type 3 and R434 Recurrent episodic apnoea were retired as they are not currently part of the Test Directory v5.
- Panel names were changed for R440 Neuropophyseal diabetes insipidus and R215 CDH1-related cancer syndrome.
- Gene content was updated for R440 Neuropophyseal diabetes insipidus, R215 CDH1-related cancer syndrome and R221 Familial tumours of the nervous system.
14th September 2023: Addition of single gene tests and small non-WGS panels
We have now added panels for the single gene tests and small non-Whole Genome Sequencing (WGS) panels for the NHS Genomic Medicine Service (GMS) that were not previously included in PanelApp. Previously these were only available in the NHS National Genomic Test Directory documentation. This has been achieved with our partners at NHS England.
In this update we have added and signed off 109 GMS panels for clinical indications in the current NHS National Genomic Test Directory v5, which include 93 single gene tests and 16 small non-WGS panels. This brings the total number of panels for the GMS from 191 to 300.
22nd March 2023: Update of GMS panels
We are pleased to announce our second update to the NHS Genomic Medicine Service (GMS) panels within 6 months. This has been achieved with our partners at NHSE and our community of reviewers.
In this update we have signed off 147 NHS GMS panels. There have been over 950 updates to the green (diagnostic level evidence) entities on the panels including the addition of new genes and entities, as well as rating and mode of inheritance changes.
In addition, the names of 41 panels have been updated to align with the associated clinical indication names in the NHS National Genomic Test Directory.
Two small panels for clinical indications in the current NHS National Genomic Test Directory v4 (R215 CDH1-related cancer syndrome and R216 Li Fraumeni Syndrome) have been added. Further small panels will be added to PanelApp in the coming months.
A further seven panels for new clinical indications have been added:
- R430 Inherited prostate cancer
- R434 Recurrent episodic apnoea
- R436 Hereditary alpha tryptasaemia
- R438 Paediatric pseudo-obstruction syndrome
- R439 Fuchs endothelial corneal dystrophy type 3
- R440 Neuropophyseal diabetes insipidus
- R441 Unexplained death in infancy and sudden unexplained death in childhood
The first 6 of these panels are associated with non-WGS tests. R441 will be tested via WGS and will be available for ordering through the GMS test ordering system in the next few weeks.
30th November 2022: New GMS panel versions signed-off
Today (30/11/2022) 134 NHS Genomic Medicine Service (GMS) panels, including 7 new panels, were updated and signed-off in PanelApp. This release includes the addition of new green genes and entities, as well as rating and mode of inheritance changes. Two panels (R151 and R152) have been merged to form a single panel, namely ‘Familial hyperparathyroidism or hypocalciuric hypercalcaemia’. The percentage overlap thresholds for regions (CNVs) have been updated from 80% to 60% to allow better detection and some have new genomic coordinates aligned with ClinGen. Finally, normal and pathogenic repeat lengths of some STRs have also been updated.
The PanelApp Handbook has been updated to describe the method for calculation of gene coverage profiles for the genes that will be green on the newly signed off panels.
Overall, this release represents over 1700 data changes to green entities across the panels and is the first major update to GMS panel content since their launch at the start of 2020.
This has been achieved with our partners at NHSE and our community of reviewers, highlighting the value of worldwide collaboration to improve scientific knowledge. Continued updates to the PanelApp knowledgebase will increase diagnostic rates as demonstrated by previous investigations at Genomics England leading to improved patient outcomes.
29th July 2021: New PanelApp publication
Read our new publication in the American Journal of Human Genetics highlighting the benefits of using the PanelApp platform in harmonising gene and panel curation efforts across healthcare systems in collaboration with PanelApp Australia.
Scaling national and international improvement in virtual gene panel curation using a collaborative approach to discordance resolution, AJHG, 2021
May 2021: NHS GMS Panels Resource Now Available!

We are pleased to announce the launch of the NHS Genomic Medicine Service (GMS) Panels Resource
, which offers a view of the 173 signed-off panels that relate to genomic tests listed in the NHS National Genomic Test Directory. This platform contains only ‘Green’ (diagnostic level of evidence) genes, STRs, and regions (CNVs) that have been approved for diagnostic testing in the NHS in England and is intended for use by NHS users and clinicians.
The main Genomics England PanelApp knowledgebase here will be dedicated for use by the wider scientific community for visualisation and reviewing of genes/genomic entities on virtual gene panels and will continue to be updated by Genomics England curators.
For more information on the evaluation and approval process of the content of GMS signed-off panels, please refer to the updating the NHS National Genomic Test Directory webpage.
Click here to go to the NHS GMS Panels Resource
5th May 2021: PanelApp has been successfully upgraded to v3.2.0
This release includes the following new features and bug fixes:
- On panel pages, links from the NHS Genomic Medicine Service (GMS) signed off panels version numbers to a view of diagnostic-grade 'green' genes/entities in the version in the NHS GMS Panels Resource have been added
- Functionality to allow listing of the latest and previous versions of panels that have been signed off for the NHS GMS on relevant panel pages
- The /api/v1/panels/signedoff/ endpoint for the API will now list the latest signed off version number of panels by default. All signed off versions can be listed when the display=all filter is used. For individual panels, all signed off version numbers can be obtained using the panel_id= and display=all filters. Note: the /api/v1/panels/signedoff/{id} endpoint will be deprecated by the end of 2021. Instead, please use the list of signed off panels endpoint with panel_id= parameter to get the latest version, and then use /api/v1/panels//?version=. to get the panel data.
- Improved validation of the names for Short Tandem Repeats (STRs) and regions, so that the correct format for names is followed.
22nd February 2021: An updated Genomic Imprinting gene panel is available in PanelApp
We have recently updated the Imprinted genes panel on PanelApp. This panel lists genes and entities that are involved in Human Genomic Imprinting and brings together information from various sources, including the Imprinting Disorders, Genomics England Clinical Interpretation Partnership (GeCIP) Subdomain (November 2015), Catalogue of Parent of Origin Effects, Geneimprint, and Tucci et al 2019 (PMID:30794780). It currently lists 231 entries and is now called Genomic imprinting, to reflect that it includes imprinted genes and others which are involved in the imprinting process but are not imprinted themselves.
We would like to encourage the Imprinting community and others to use this resource, not only to have easy access to the information, but also to add value to it by the submission of additional genes and reviews of the existing genes listed on the panel using PanelApp’s crowdsourcing features. If you would like to add genes or add comments to existing genes please register as a reviewer.
21st August 2020: PanelApp reaches 5 years old!
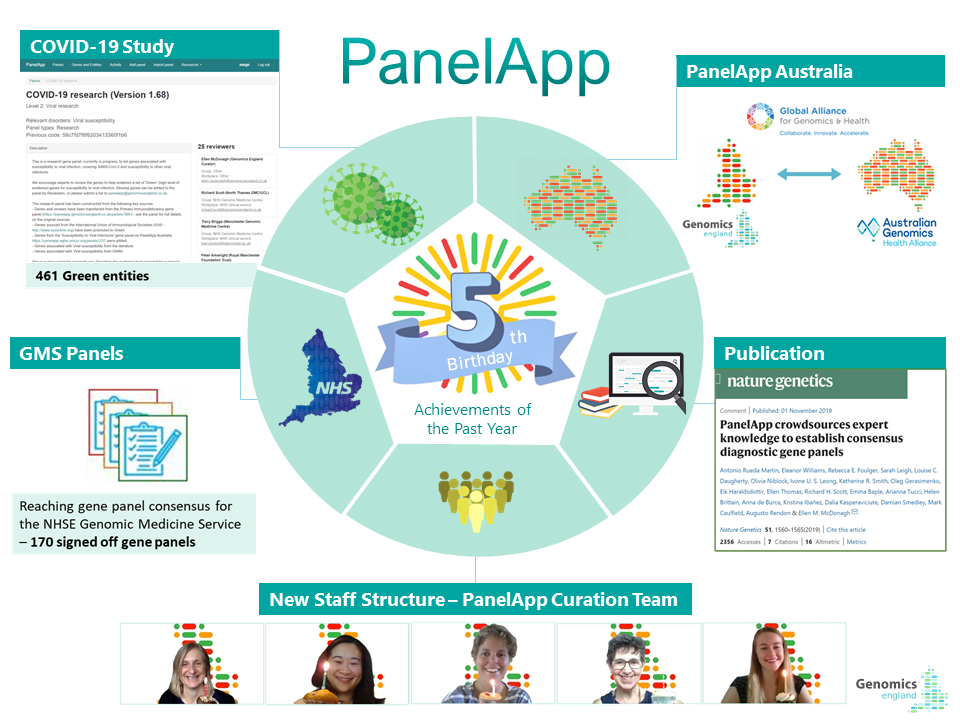
Today we are celebrating PanelApp’s 5th birthday! Looking back over the year, we are very proud of all that we have achieved. Here are five of our highlights:
1. PanelApp Publication – November 2019 saw the publication of our PanelApp paper in Nature Genetics. The paper informs the international genetics community about PanelApp and its importance in the community. It also allows users to acknowledge the contribution of PanelApp to their work by citing this publication in their material.
2. PanelApp became International - PanelApp Australia was launched in December 2019, thanks to the dedicated work of Zornitza Stark and colleagues. We are working to complete comparisons of panels between the two resources.
3. COVID-19 Research Panel - As part of Genomics England’s contribution to the GenOMICC consortium, the PanelApp curation team created a COVID-19 Research gene panel, which will be used in some analyses of patients who have had COVID-19. The panel creation was a truly collaborative effort within the community, with amazing contributions from the Illumina clinical curation team and researchers and reviewers from all over the world.
4. Consensus GMS Panels - PanelApp was used as the platform and along with the skills of the curation team, allowed for consensus gene panels for the NHS England Genomic Medicine Service, resulting in 170 GMS signed-off gene panels to be made. The Green genes, STRs and Regions on these panels will be covered by the tests in the National Genomic Test Directory as a national consensus of which entities are tested for each clinical indication.
5. PanelApp Curation Team - We sadly said goodbye to some members of the curation team; Ellie McDonagh, the founder of PanelApp and the Genomics England curation team, along with Louise and Rebecca. Their contributions were invaluable in making PanelApp the successful product it is today, and we wish them all the best for the future. It means we have been joined by Arina who is already making fantastic contributions to PanelApp.
None of this would be possible without the support and contributions we receive from curators, developers, clinicians, labs, reviewers, collaborators, participants & users from around the world.
A big thank you to all.
27th July 2020: PanelApp has been successfully upgraded to v3.1.2
This release includes the following new features:
-
The "Download signed off version" button will appear at the top of the panel page for Genomic Medicine Service panels, allowing users to easily see the ability to download the 'GMS-signed off version' of a panel.
-
The blue banner at the top of GMS-signed off panels has changed the wording to “Version {number} of this panel was signed-off for the GMS. The current version, shown here, may differ from the signed-off version”. This is to add clarity between the actual version of the panel and GMS-signed off version of panels.
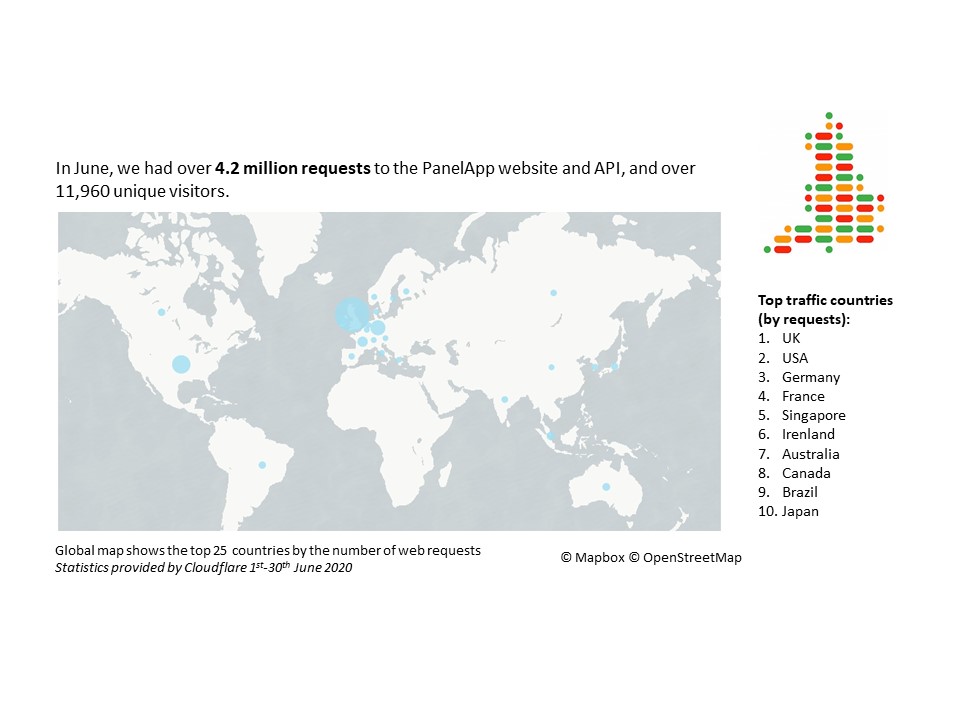
June 2020 statistics
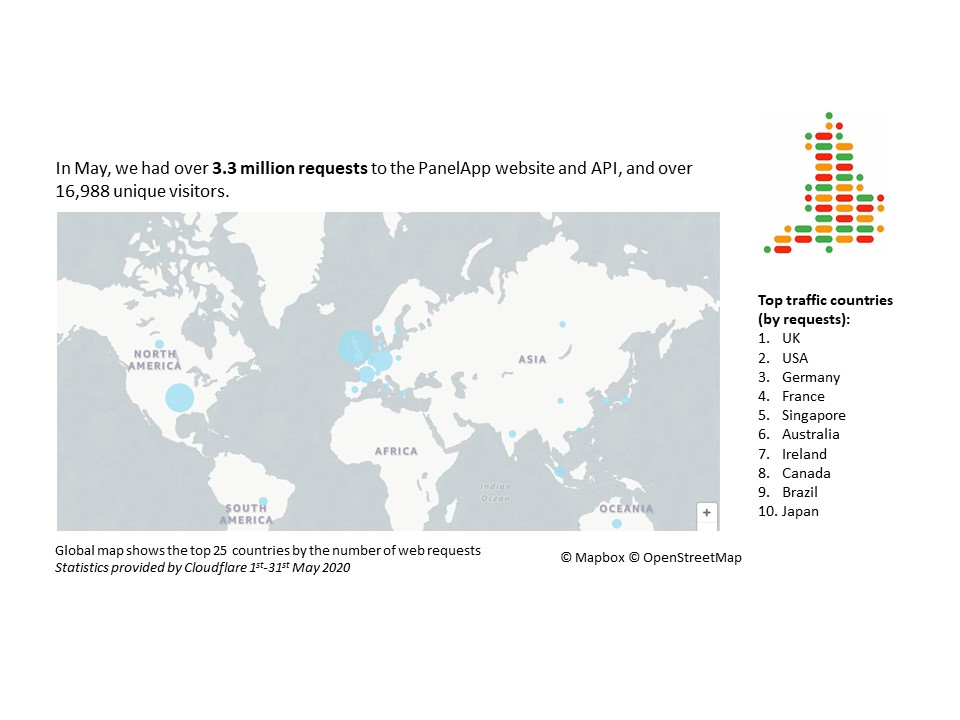
May 2020 statistics
The Genomics England Covid-19 version 1.0 research gene panel
We are delighted to announce that the Covid-19 panel (https://panelapp.genomicsengland.co.uk/panels/111/) has be created and is now version 1.0, ready to be used for analysis.
The Genomics England Curation team would like to thank all of those involved in this fast-paced endeavour to create a research panel, including Expert reviewers, Illumina and PanelApp Australia (https://panelapp.agha.umccr.org/).
You can still contribute to the Covid19 panel content by adding genes or expert reviews at: http://panelapp.genomicsengland.co.uk/panels/111/
PanelApp Journal Review
On Thursday 4th June, the curation teams in both Genomics England and PanelApp Australia will be reviewing publications in a wide variety of current journal editions for novel gene-disease discoveries and additional case reports or functional studies relevant to our rare disease panels. Want to join the effort? If you have recently read or published a paper relating to rare disease, let us know. You can register as an Expert Reviewer in PanelApp to add or review genes directly on a panel. Alternatively contact us by email.

Can you review our curated COVID-19 research gene panel in PanelApp?
Genomics England is partnering with the GenOMICC consortium, Illumina and the NHS to sequence human genomes in the fight against coronavirus. As part of this project, we have created a PanelApp research panel to identify human genes linked to susceptibility to viral infection. The panel has a broad scope and covers
susceptibility to SARS-CoV-2 and other viruses. This will aid research into whether host genetic determinants could explain at least part of the severity of a COVID-19 infection. The Description box of the panel provides a brief summary on how the panel has been constructed so far. You can register as an Expert Reviewer in PanelApp to add or review genes directly on the panel itself. Alternatively contact us on [email protected] with additional gene lists. Thank you for your contribution to the project!
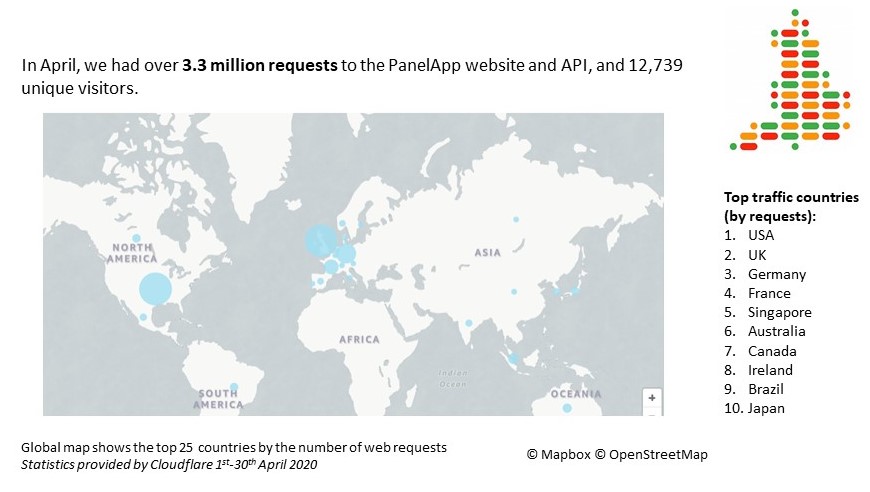
April 2020 statistics
Star Reviewer: Dr Helen Brittain

Recently we began a series of Star Reviewer posts, highlighting experts and community members who have made a significant contribution to PanelApp. Our recent blog focuses on Dr Helen Brittain:
Dr Helen Brittain is a consultant clinical geneticist at Birmingham Women’s and Children’s NHS Trust, and a senior clinical fellow in rare disease genomics at Genomics England. Helen qualified in medicine from the University of Nottingham, before training in Paediatrics and then undertaking clinical genetics training in London, based at Great Ormond Street Hospital. She has a particular interest in paediatric developmental disorders.
Helen provides regular clinical input to the Genomics England curation team, answering weekly queries on panel scope, phenotypic relevance and any other clinical questions. Her breadth of knowledge helps us curate across our 325 PanelApp panels. Not withstanding her additional Genomics England clinical work, Helen has made an incredible contribution to PanelApp. Since January 2017, Helen has contributed over 1000 direct evaluations to PanelApp for 56 panels spanning Neurology, Paediatrics, Cancer susceptibility, Endocrinology, Dermatology, Skeletal disorders, Cardiac disorders (and that's just a few). Helen was also recently our principal advisor on the creation of the individual ciliopathy panels and the multisystem ciliopathy panels for use in the NHS Genomic Medicine Service (e.g. https://panelapp.genomicsengland.co.uk/panels/728/).
Thank you so much Helen for your continued reviewing and clinical advice, from all the Genomics England Curation Team.
Viral susceptibility panel
We have created a PanelApp research panel to capture the genes involved in viral infection and susceptibility. Please contribute by registering as an Expert Reviewer and adding and/or reviewing human genes linked to viral infection. The Viral susceptibility panel can be found at: https://panelapp.genomicsengland.co.uk/panels/111/
March 2020 statistics
![PanelApp_Usage]
(https://panelapp.genomicsengland.co.uk/media/images/PanelApp_stats_March2020crop.jpg)
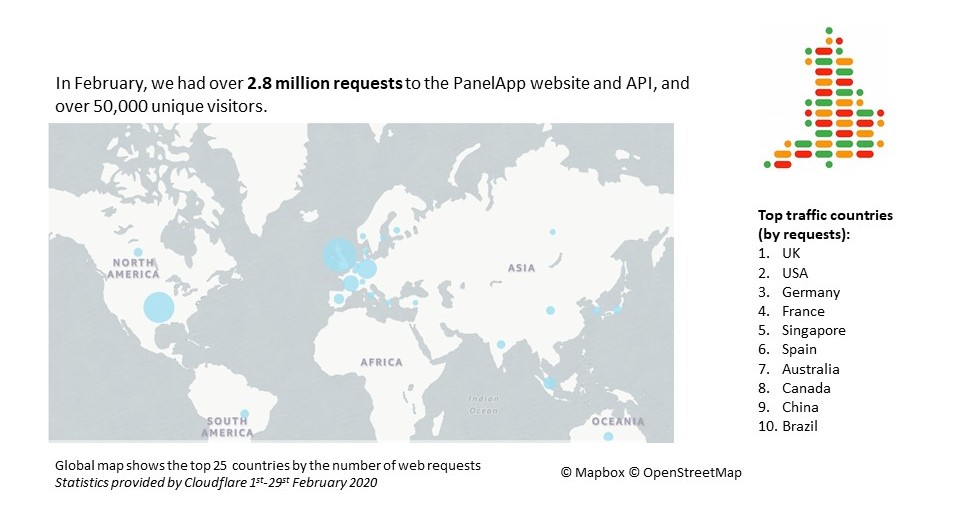
February 2020 statistics
Rare Disease Day 2020

Rare Disease day this year falls on the most rare of dates: February 29th. PanelApp is pleased to be able to support Rare Disease patients and their families with high-quality, manually-curated gene panels for interpretation of rare diseases. The panels have been manually curated by Genomics England curation team through collaborations with experts in the NHS and from around the world including clinicians, clinical scientists and researchers. The panels are free to browse and download from PanelApp.
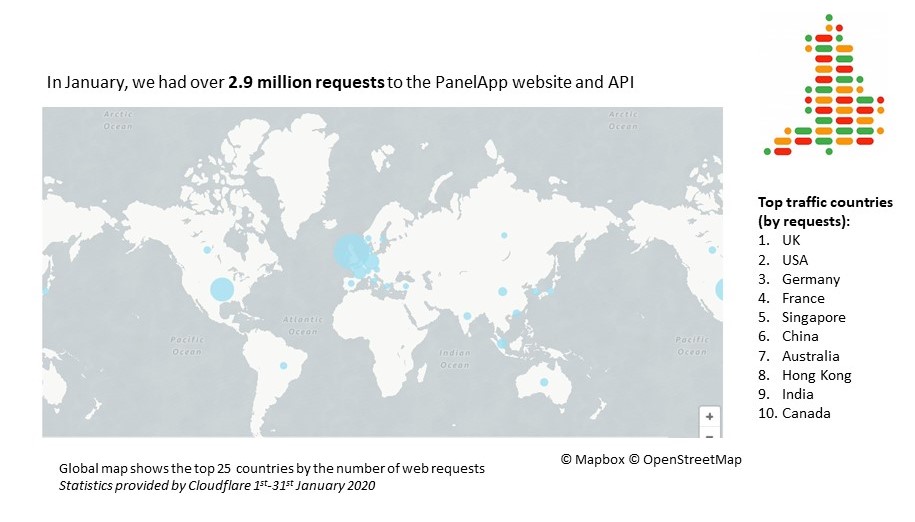
January 2020 statistics
5th Feb 2020: PanelApp has been successfully upgraded to v3.1.1.
This release includes:
- changes to the signed off panel API endpoint to display panel count and pagination
27th Jan 2020: PanelApp has been successfully upgraded to v3.1.0.
This release includes:
- New fields allowing capture of information for the Genomic Medicine Service
- Display of super panels on the gene/entity page. E.g. ADAR
- Optional transcript field E.g. APOB
- Renaming of the homepage tab 'webservices' to 'API'
- Additional fixes to uploads to standardise 'Mode of Inheritance' terms.
- Performance optimisation
21st Jan 2020 Panel Swap!
You may have noticed PanelApp Australia panels appearing in Genomics England PanelApp and vice versa over the last few weeks! We are swapping panels to enable systematic comparison, starting with renal disease, epilepsy, intellectual disability and hearing loss panels.
Through this process, we hope to build consensus internationally and improve the evidence base for diagnostic practice in rare disease genomics. The PanelApp Australia panels can be identified in Genomics England PanelApp by filtering for ‘VCGS’ (Victorian Clinical Genetics Services) in the panel name. These panels also have Panel Type ‘External Diagnostic Lab’.
To view the original VCGS panels, visit the PanelApp Australia website.
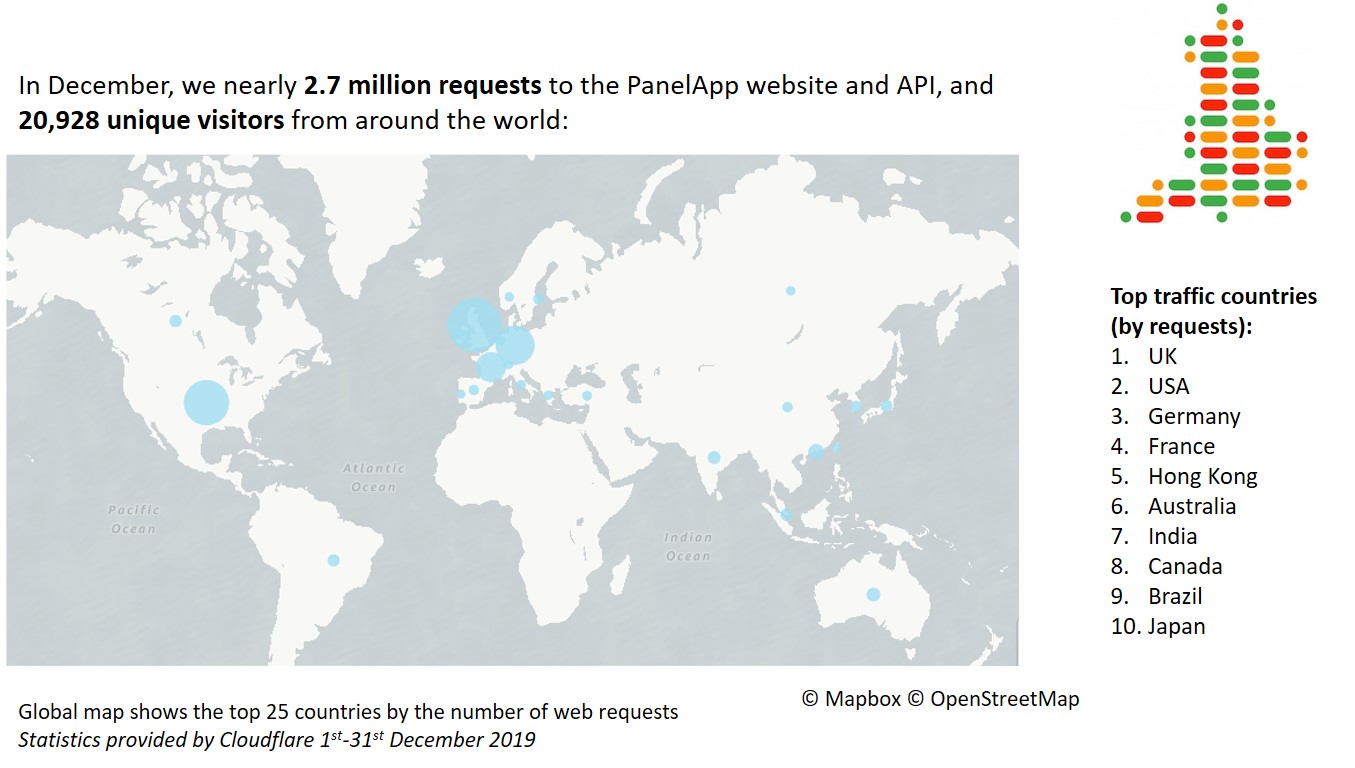
December 2019 statistics
17th Dec 2019 Australian Genomics launches local instance of PanelApp
Earlier this year, we made the Genomics England PanelApp software open source, allowing others to utilise the code for the crowdsourcing and curation tools we have developed for their own endeavours.
Currently in Australia, the process for documenting and sharing gene-disease associations is manual and inefficient. An Australian instance of PanelApp has been launched by Australian Genomics, allowing the consolidation of multiple disparate silos of activity into a single open national platform will reduce the gene curation burden on individual laboratory and clinical services and improve diagnostic outcomes for Australian patients.
“PanelApp Australia marks a huge step forward in enabling more efficient and robust diagnostic practices in Australia. I encourage local laboratories, clinicians and researchers to sign on to the platform and lend their expertise”
- Associate Professor Zornitza Stark from Australian Genomics, coordinator of the content management for the platform.
The teams at Genomics England and Australia Genomics plan to continue to collaborate, and are hoping to develop ways to integrate the two instances that will allow sharing of gene-disease curation. Read more on the Genomics England website here.
16th December 2019: Spectrum news article
PanelApp features in a Spectrum article regarding useful resources for genes and variants for autism research. In PanelApp, we have set up a research gene panel for autism and are seeking experts to review the genes on this panel.
Easy steps to add your expert review:
-
Please register to be a reviewer here
-
Once we approve your account, log in and go to the research gene panel for autism
-
Click on a gene. You will see the review tool - provide your expertise by adding a rating for the gene and any further evidence such as publication or comments.
Your name and affiliation will appear on the panel in acknowledgement of your contribution. Thank you! Your review will contribute to establishing a consensus gene panel for genes with evidence for causation in autism.
More details on being a reviewer are provided in the PanelApp Reviewer's Guide
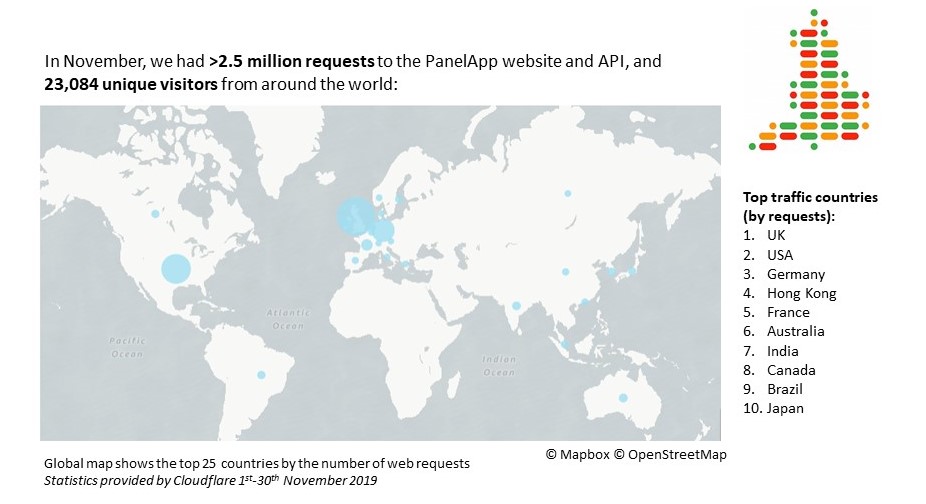
November statistics
1st November 2019: PanelApp publication out!
Nat Genet (2019) doi:10.1038/s41588-019-0528-2
Antonio Rueda Martin and Eleanor Williams, Rebecca E. Foulger, Sarah Leigh, Louise C. Daugherty, Olivia Niblock, Ivone U. S. Leong, Katherine R. Smith, Oleg Gerasimenko, Eik Haraldsdottir, Ellen Thomas, Richard H. Scott, Emma Baple, Arianna Tucci, Helen Brittain, Anna de Burca, Kristina Ibañez, Dalia Kasperaviciute, Damian Smedley, Mark Caulfield, Augusto Rendon & Ellen M. McDonagh
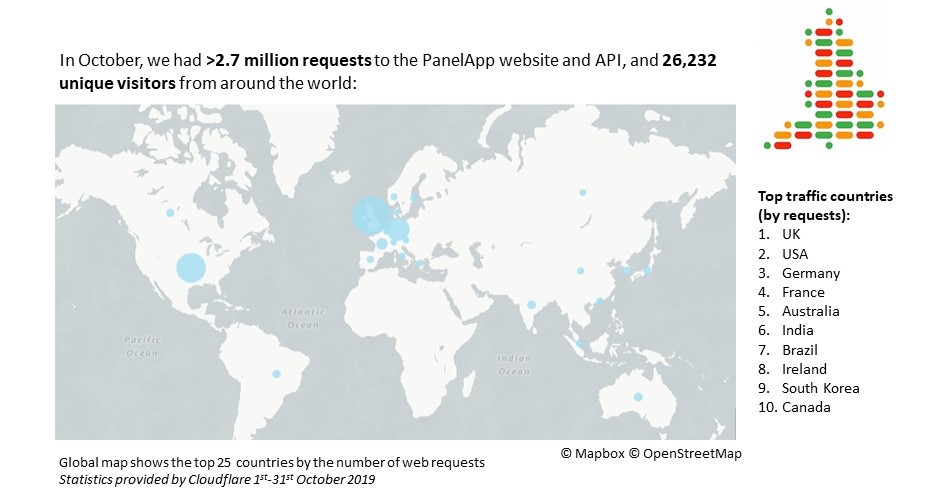
October statistics
Star Reviewer: A/Prof. Zornitza Stark
PanelApp has a crowdsourcing tool to allow each gene to be reviewed and commented on by experts within the scientific community. This helps gain a consensus on which genes have a diagnostic-grade level of evidence for association with a disease, and ultimately impacts on patient diagnosis. PanelApp has nearly 900 reviewers registered, bringing together a community of experts with different backgrounds from around the world. Expert review plays a huge part in ensuring that the PanelApp gene:disease associations reflect the latest knowledge. To acknowledge the contribution and scale of this input, we will be posting a series of blog entries highlighting some of our Star Reviewers, beginning with Zornitza Stark

A/Prof. Zornitza Stark is a clinical geneticist at the Victorian Clinical Genetics Services (VCGS) in Melbourne, and clinical research fellow with Australian Genomics. She completed her medical studies at the University of Oxford, before training in paediatrics at the Royal Children’s Hospital in Melbourne, and in clinical genetics at VCGS. She was awarded a doctorate in clinical genomics from the University of Oxford in 2017, and in 2018, Zornitza spent a short sabbatical at Genomics England.
Zornitza Stark has made phenomenal contribution to PanelApp over the past year. Initially, she spent a number of weeks with us at Genomics England in London becoming familiar with PanelApp. She shared the lists of genes that were being used by the Victorian Clinical Genetics Services and Australian Genomics, and started to use her considerable clinical genetic expertise to review genes on our panels. Since returning to Australia she has continued to review genes and to add more genes to PanelApp where appropriate. Her reviews are detailed, including up to date publications, information about unpublished cases and her clinical opinion about the relevance of a gene to a particular phenotype. She has contributed over 400 reviews, and her expertise has allowed well over a hundred different genes to be rated on over twenty five panels, including Genetic epilepsy syndromes, Intellectual disability and Mitochondrial disorders. This considerable achievement sets her out as a PanelApp Star Reviewer.
Thank you very much Zornitza, from the Genomics England Curation team.
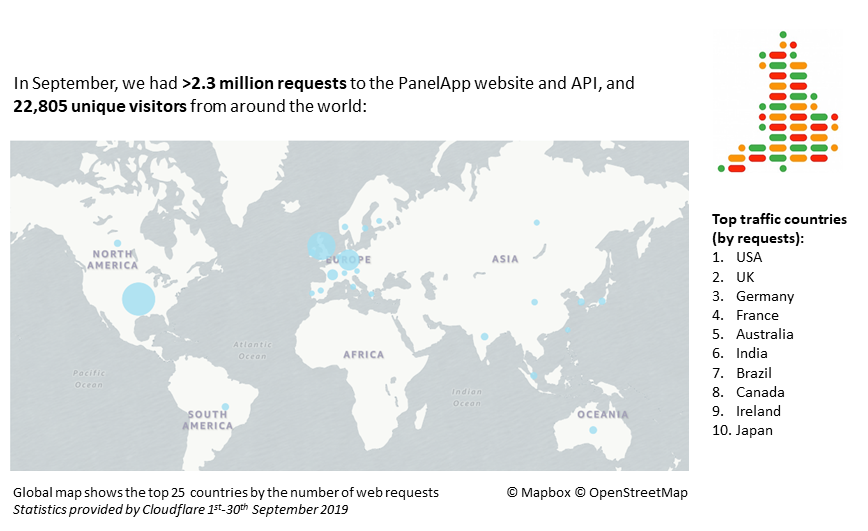
September statistics
23.09.2019 New PanelApp Handbook
We have updated our PanelApp Handbook to V7.0 to guide you through all our new features, including:
-
New panel types, including Super panels
-
How to find panels for the Genome Medicine Service (GMS)
-
Effectively querying the API
-
PanelApp 2.4.2 features
A Fetal anomalies Virtual Gene Panel for the Genomic Medicine Service.
Anna de Burca, Genomics England Clinical Fellow, will be presenting at the 23rd International Conference on Prenatal Diagnosis and Therapy in Singapore on 7-11 September.
Dr de Burca will be giving a Lightning Communication and presenting a poster on how the PanelApp curation platform was used to develop the Fetal anomalies gene panel for the NHS England Genomic Medicine Service.
The finalised panel will be used to interpret rapid exome sequencing results for fetuses with multiple abnormalities where a monogenic malformation disorder is considered the likely aetiology. The panel contains genes with robust gene-disease associations where the phenotype is fetally-relevant, and was created in collaboration with colleagues at Great Ormond Street Hospital.
A preview of Dr de Burca's poster is available here, and you can follow the conference using #ispd2019
August statistics:

21st August 2019: Today is PanelApp's 4th Birthday!
PanelApp was launched 4 years ago. Today we reflect on some of our achievements, and would like to thank everyone who has contributed to PanelApp over the years - this includes our Curators, Developers, Bioinformatics and Clinical Team within Genomics England, Expert Reviewers from around the world, Participants of the 100,000 Genomes Project, collaborators and those who have helped promote PanelApp or utilise PanelApp.
Here's a summary of a few of our achievements in the last 4 years since Panelapp was launched:
-
We now have 56,640 gene-disease curations.
-
There are 310 public gene panels available to review, browse, query.
-
There are 3139 unique Green (diagnostic-grade) genes
-
We expanded the scope of PanelApp beyond genes with the addition of Copy number variants (CNVs) and Short Tandem Repeats (STRs) implicated in rare diseasesm so they could be added to our genome interpretation pipeline. We now have 18 unique Green (clinically-relevant) STRs on 27 different gene panels and 62 unique Green (clinically-relevant) CNVs on 60 different gene panels.
-
Our curated version 1+ gene panels cover all the rare diseases relevant to participants recruited in the 100,000 Genomes Project, covering around 50% of OMIM phenotypes. We continue to update these gene panels with new genes as evidence arises, and encourage people to add their reviews.
-
We have incorporated PanelApp into the Genomics England cancer germline genome analysis, providing 21 different gene panels for pertinent cancer susceptibility.
-
PanelApp is utilised globally, with around 2 million requests to our website and API, and over 10,000 unique visitors worldwide per month, contributing to research and other clinical projects.
-
PanelApp is a member of the Gene Curation Coalition, with other key databases and curation endeavours such as OMIM, ClinGen, Gene2Phenotype, Orphanet.
-
PanelApp data or has links to PanelApp incorporated within key databases including DECIPHER, Open Targets, Varsome, DisGeNET, Mutalyser/Mutation Taster, RDConnect. PanelApp links to other databases such as OMIM, ClinVar and Gene2Phenotype.
-
PanelApp is accredited under the Genomics England ISO 15189 Accreditation Schedule.
-
PanelApp was endorsed by the UK Genetic Testing Network (UKGTN) in 2016, and is now being used as the platform for gaining consensus gene panels for the NHSE Genomic Medicine Service.
-
In November 2017 our release supported both GRCh38 and GRCh37 genome builds.
-
Earlier this year we made the PanelApp software code Open Source.
-
We deployed PanelApp to Amazon Web Services Native Cloud yesterday
20th August 2019: PanelApp has successfully been upgraded to v3.0.0 and is now on Amazon Web Services Native Cloud.
What does this mean?:
-
Faster response times and better availability
-
New features and improvements for PanelApp can be released quicker, due to streamlined development and deployment processes (CI/CD)
-
Enhanced security
July statistics
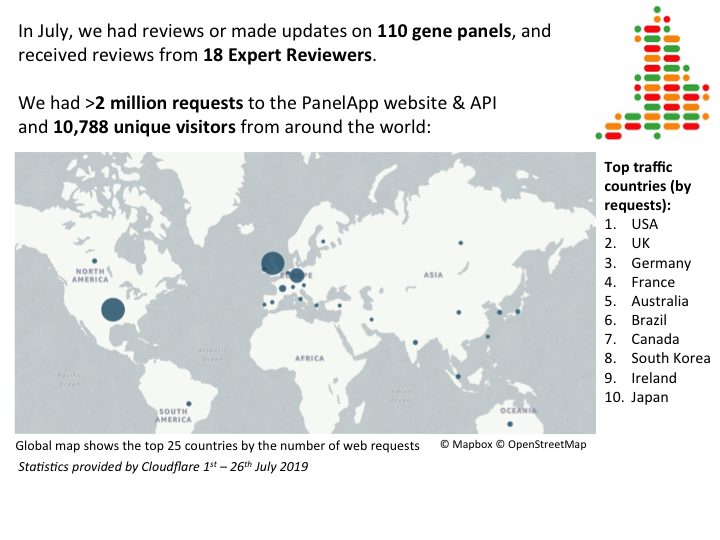
31th July 2019: ACGS conference 2019 summary
In June, Dr Ivone Leong from the PanelApp Team attended the Association for Clinical Genomic Science (ACGS) Summer Meeting 2019 in Birmingham, UK. The meeting was held for two days and the clinical genetics community from around the United Kingdom gathered and presented talks and posters on topics ranging from new testing techniques, new variant/gene discoveries and interesting case studies.
Ivone presented on “A process for gaining consensus gene panels in the Genomic Medicine Service using PanelApp”, which explained how PanelApp is being used as the repository for defining the content of gene panels that will be used within the NHS Genomic Medicine Service (GMS). The process of reaching consensus gene panels for the GMS was outlined and the role of the PanelApp Curation Team within this. Ivone also raised the challenges the PanelApp Curation Team have encountered and overcome, and the overall benefits PanelApp has gained through these experiences.
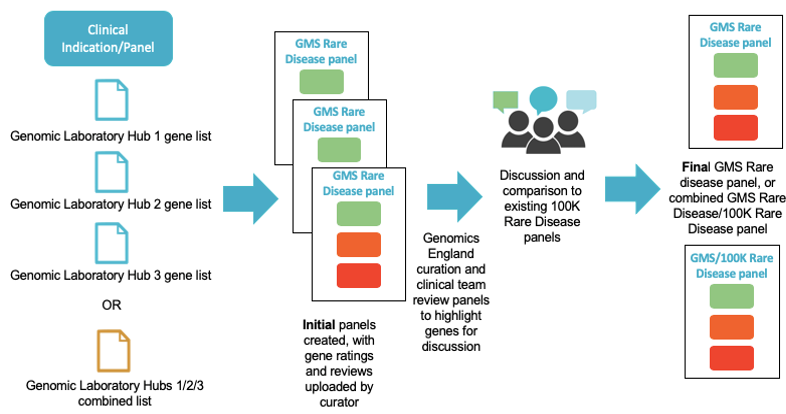
29th July 2019: PanelApp upgrade to v2.4.2
Please note: PanelApp will be upgraded to v2.4.2 at 10am (BST) on Monday 29th July for a minor fix. Please note that users of Panelapp should not experience any significant downtime for this upgrade, it may be down for up to one minute.
1st July 2019: PanelApp Web Requests
PanelApp Reviewers and Users are worldwide, allowing PanelApp to have expert input from across the globe. It's encouraging to see a wide distribution of PanelApp users between countries. Below is a map detailing PanelApp web requests from June 2019, with the top 25 countries shown on the right hand side.
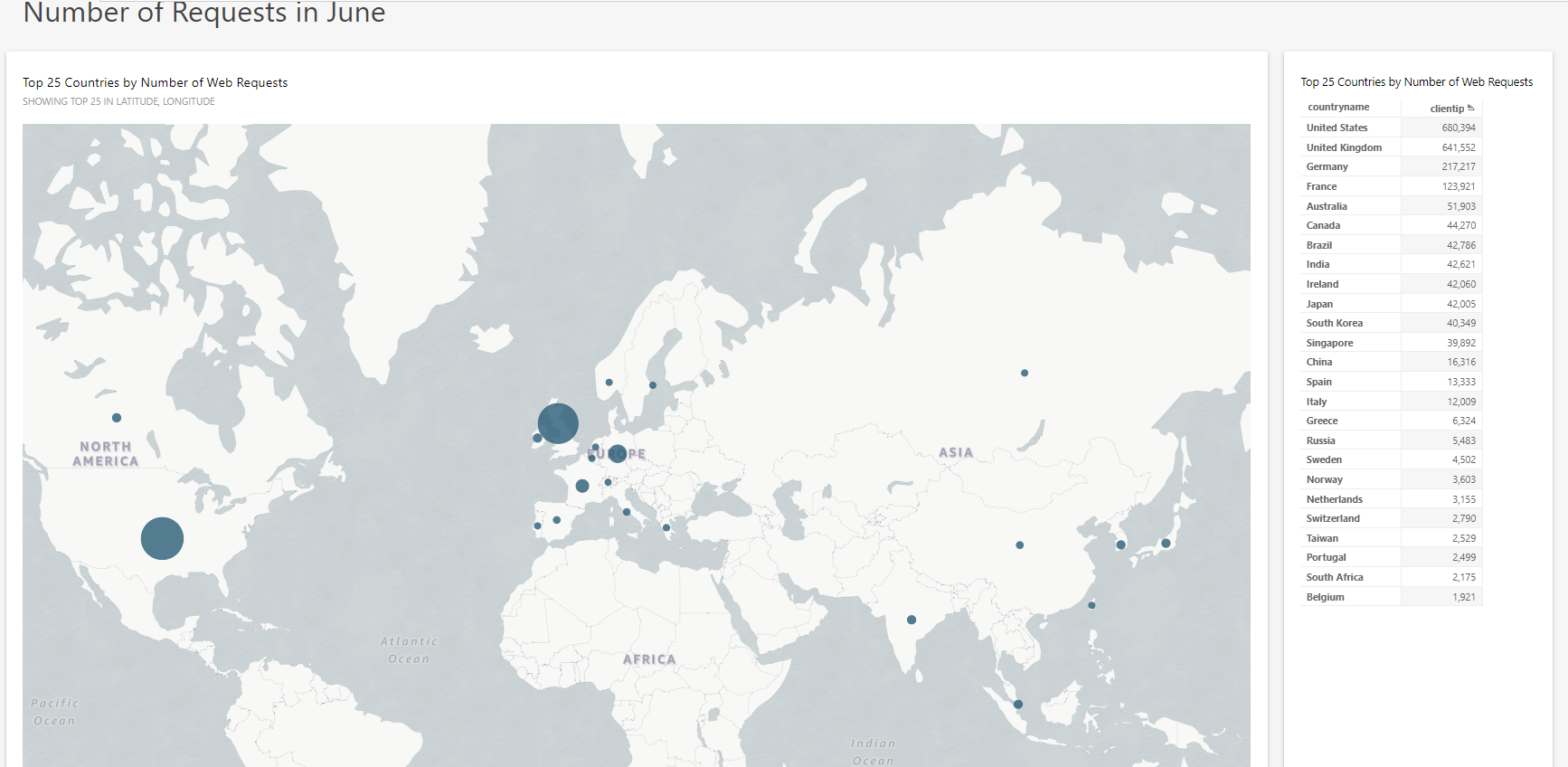
When the requests are corrected for number of unique hits- United States, China and the UK come out top. Let us know if you have any questions about PanelApp as you browse our tool.
20th June 2019: New release of PanelApp!
PanelApp V2.4.0 has several changes to the backend to improve usability and several new features.
The 'Panel Types' field now appears on the panels page, allowing users to filter the list of panels based on this category by typing key words into the filter box (as pictured). The panel type of a gene panel describes what the panel is used for.
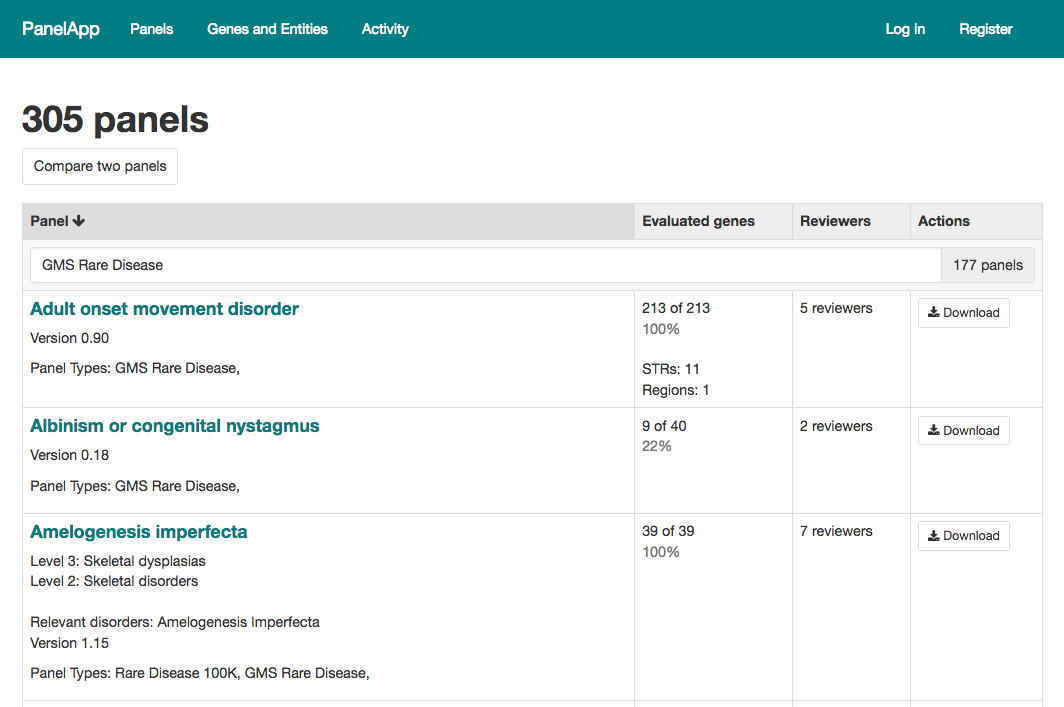
Panel types:
-
Cancer Germline 100K: a panel used for the interpretation pipeline for cancer genomes from the 100,000 Genomes Project. Example: Breast cancer pertinent cancer susceptibility
-
Rare Disease 100K: a panel used for the interpretation pipeline for rare disease genomes from the 100,000 Genomes Project. Examples: Neonatal cholestasis
-
GMS Rare Disease: a panel developed for the NHS Genomic Medicine Service - may be delivered by whole exome sequencing, large or small 'wet lab' panel or as a virtual panel. These are currently in review by NHS Genomic Medicine Service specialist groups. Example: Adult onset movement disorder
-
GMS Rare Disease Virtual: a panel developed for the NHS Genomic Medicine Service - will be used as a virtual panel for whole sequencing indications. These are currently in review by NHS Genomic Medicine Service specialist groups. Example: Hereditary neuropathy
-
Super Panel: a panel made up of component panels. When the component panels are updated, the genes on the Super panel are automatically updated. Example: Paediatric disorders
-
Component Of Super Panel: a panel that has been added to a super panel. Changes to this panel will automatically be updates on the super panel. Example: Arthrogryposis
-
Actionable: a panel containing actionable-type information linked to genes, such as clinical trial information. Example: Gene therapy clinical trials
-
Research: a panel from a research project. Example: Autism
Several new sources have also been added to the drop down menu for adding genes, including ClinGen and URLs in the description box for a gene panel are now active.
10th-11th June: ACGS conference 2019
Our Scientific Curator Dr Ivone Leong presented at the Association for Clinical Genomic Science - ACGS conference. Her talk was entitled “A process for gaining consensus gene panels in the Genomic Medicine Service (GMS) using PanelApp”, explaining how PanelApp is being used as the platform for defining the content of gene panels for the NHS GMS.
View the gene panels for the GMS that are currently under review by typing 'GMS' in the panels filter box here.
29th May - 31st May: Curating the Clinical Genome 2019
Our Head of Curation & Pharmacogenomics, Ellen (Ellie) McDonagh, was invited to speak at the Curating the Clinical Genome conference in Washington DC USA. The conference was co-hosted by ClinGen and DECIPHER, with a jam-packed schedule of everything involved in curating genomes for clinical interpretation. Presentations ranged from variant classification and reinterpretation, studying penetrance, polygenic scoring for disease prediction, to the essential need for in-depth phenotyping, data sharing, standards and partnerships.
Ellie presented within the Gene Curation session, which included other speakers on the subjects of crowdsourcing and global gene-disease curation efforts. Ellie’s talk, ”Gene Curation for the Interpretation of Clinical Genomes and the Future of PanelApp” covered the role of PanelApp in the interpretation of genomes for rare disease patients in the 100,000 Genomes Project, PanelApp as a platform for gaining consensus on gene panels for the NHSE Genomic Medicine Service, and the ongoing global collaborations we have for sharing of curation efforts and data. These include the GA4GH and Gene Curation Coalition (GenCC)…Marina DiStefano from Harvard Medical School went on to speak in more detail about GenCC activities later in the session.
Later that day, Ellie visited George Washington University Cancer Centre and Department of Biochemistry and Molecular Medicine, hosted by Dr Raja Mazumder. Ellie gave a presentation including an overview of the 100,000 Genomes Project and current curation activities at Genomics England, including PanelApp and pharmacogenomics.
Collaborating with the Curation Community at the 12th International Biocuration Conference, April 7-10th 2019.
At the start of April, Dr Ellen McDonagh and Dr Rebecca Foulger from the PanelApp Team attended the 12th International Biocuration Conference in Cambridge, UK. The conference provides a forum for curators, developers and group leaders from Industry, Academia and Healthcare organisations to collaborate and discuss all aspects of curation. An overview of the conference content can be gained through the Twitter hashtag #biocuration2019, and Marina DiStefano from the ClinGen Resource wrote an excellent summary of the conference here.
The Gene Curation Coalition (GenCC) is a group of initiatives that curate links between genes and disease, and the evidence underlying these associations. The collaboration was established to harmonize the approach to gene-disease validity curation and ensure inter-operability between different curation resources. The GenCC was represented at the conference by Genomics England PanelApp’s Ellen McDonagh and Rebecca Foulger, Marina DiStefano from ClinGen, and Antoine Marmignon from Orphanet. Together, we hosted a conference workshop to demonstrate how we collate and assess evidence behind gene:disease associations, a complex task that is critical for the accurate interpretation of genomic variants in patients.
Each group in the coalition has a different focus with a different set of users, and we began the workshop with PanelApp, ClinGen and Orphanet presenting an overview of our gene curation strategies, together with our curation tools and the webpages displaying the results. A guided browsing session on PanelApp then allowed attendees to familiarise themselves with the traffic-light rating system and the underlying data.
As hoped, the workshop was the ideal interactive setting. With over fifty attendees, the discussions launched from the very start and continued throughout the workshop - so much so that we could have continued the activities long beyond the two hours available. The discussions reinforced some of the challenges we face in performing high quality manual curation, and the rules we have in place to ensure consistent annotation. We were also able to ascertain where we could most benefit from incorporating additional ontologies and mappings into our resources, and areas that required further clarity; it quickly became apparent that even the term 'panel' can be ambiguous given its different use between resources - are we talking about a panel of genes, or a panel of experts?
One of the key roles of the GenCC is to standardise terms used in curation, and the workshop concluded with Marina polling workshop attendees for their views on which terms should be adopted. Curators are already familiar with the need for consistent curation terminology, and Marina’s summary of the GenCC efforts to reach a set of consensus terms for validating gene:disease associations emphasises how we are already working together to allow efficient data sharing.
09.04.2019 Genomics England PanelApp software is now open source!
In her Keynote talk at the Biocuration 2019 conference, our Head of Curation & Pharmacogenomics Dr Ellen McDonagh announced that the PanelApp software code is now open source and available in GitHub.
The software behind Genomics England’s PanelApp, a crowdsourcing platform for sharing and evaluating gene panels, has now been made publicly available for the scientific and clinical community to use.
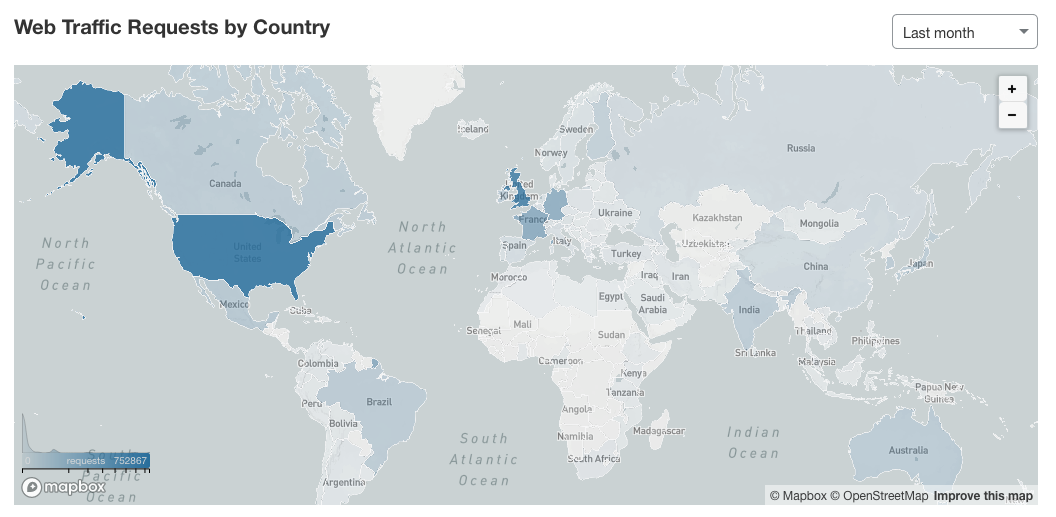
PanelApp is a knowledgebase of virtual gene panels for rare diseases and cancer. It has a crowdsourcing tool that allows experts from around the world to provide reviews of genes and the underlying evidence for disease causation (Figure 1). These panels are publicly available to browse, download, and query. In the last month there were more than 2 million requests on the PanelApp website and application programming interface (API), and more than 10,500 unique visitors from around the world (Figure 2).
Figure 1: Review tool
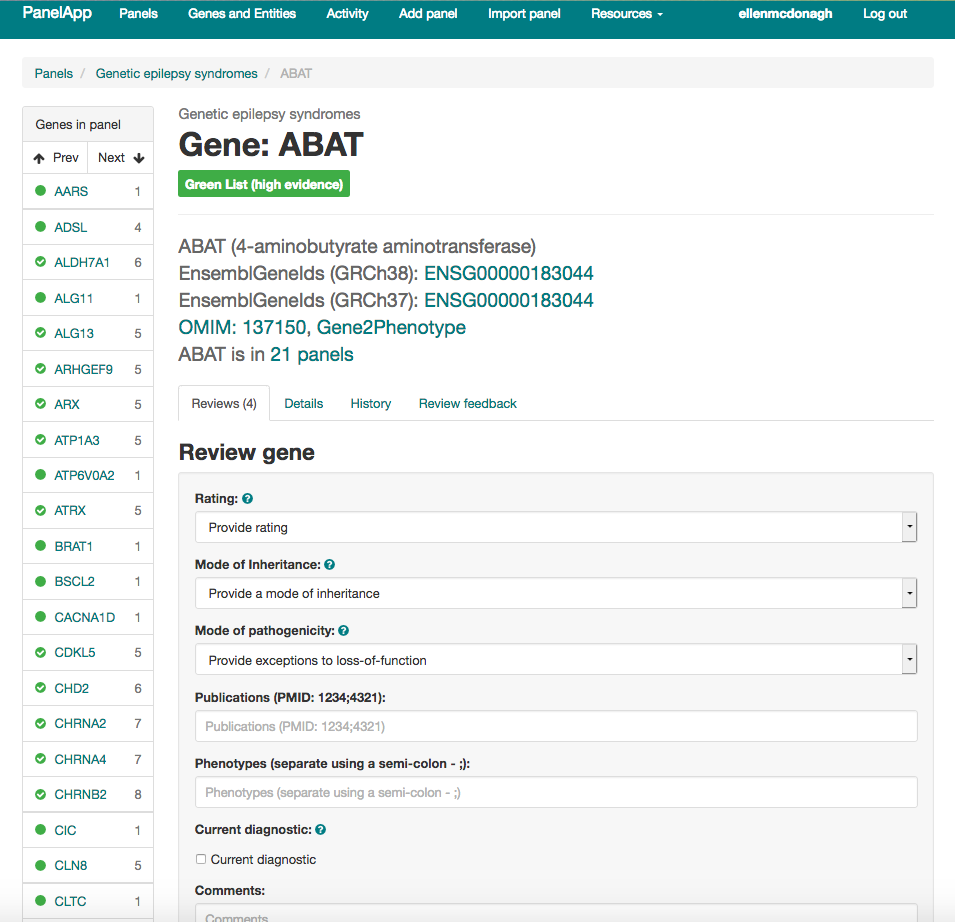
Figure 2: map of PanelApp users in the last month
We have now released the underlying code for the tools and functionality of PanelApp in GitHub.
The gene panels are used in the Genomics England interpretation pipelines for the 100,000 Genomes Project to help prioritise variants to gain diagnoses for rare disease patients, or highlight important inherited variants in cancer susceptibility genes for cancer patients. PanelApp is also being used as the platform for achieving consensus panels for the NHS Genomic Medicine Service. In each gene panel, genes are ranked using a traffic light system. The Genomics England Curation Team assess crowdsourced expert reviews and evidence from other sources to define which genes have a high level of evidence for causation in a disease. Gene panels with strong evidence are classified as “Green” for genome interpretation (Figure 3).
Figure 3: traffic light system for evidence level of gene-disease
We have had interest in PanelApp from national genomic endeavours in other countries, who are now able to use the code to install their own instance of PanelApp. As part of the Global Alliance for Genomes & Health (GA4GH) initiative, we hope to collaboratively develop a shared API to allow knowledge sharing between instances of PanelApp.
A big thank you goes to Antonio Rueda and Oleg Gerasimenko in the Genomics England Bioinformatics Team who have built and developed the PanelApp software. We would also like to thank Paul Hayes who contributed to software development, and members of the Curation and Clinical Teams for input into the tools and functionality.
We would love to hear from you! Please let us know by email: [email protected] or Twitter if you are using PanelApp in your research or diagnostic lab, or plan to install your own instance of PanelApp, as we are always interested in knowing how PanelApp is being used and how we can make improvements.
01.04.2019 PanelApp is heading to the 12th International Biocuration Conference Next Week
Ellen McDonagh and Rebecca Foulger from the PanelApp curation team will be attending the 12th International Biocuration Conference next week, being held in Cambridge UK from 7-10 April 2019. We have a busy week, starting with co-presenting a workshop with ClinGen and Orphanet on curating gene:disease associations. Ellen is an invited speaker, and will be giving a plenary talk on Tuesday 9th in the 'Curation for human health and nutrition' session. Rebecca is co-chairing this session of talks, and presenting a poster on PanelApp and our Curation Collaborations on Tuesday evening. You can follow the conference on Twitter by following #biocuration2019.
29.03.2019 PanelApp update of user statistics
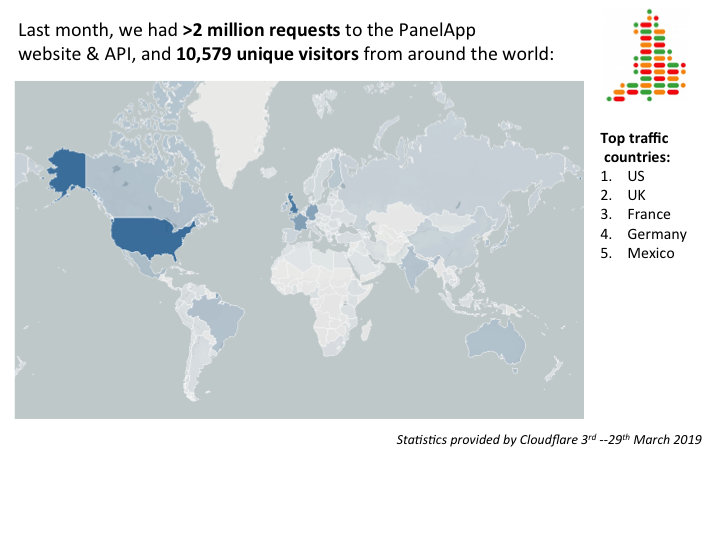
27.03.2019 PanelApp is at the Genomics of Rare Disease conference
Eleanor Williams (PanelApp curator) and Anna De Burca (Rare Disease clinical team) presented their poster "PanelApp as a platform for achieving consensus panels for a national Genomic Medicine Service" at the Genomics of Rare Disease conference at the Wellcome Trust Genome Campus, near Cambridge, UK. There was lots of interest in PanelApp at the poster session both from people who use the 100,000 Genomes Project gene panels and from those that would like to use the PanelApp software for their own gene panels.
29.01.2019 PanelApp is upgraded from to v2.3.4
The new release includes improvements and additions to API endpoints for genomic entities, as well as improvements to panel display and performance.
11.01.2019 A Reviewer's Guide To PanelApp
We have a new handy guide for reviewers of PanelApp panels. This short presentation will guide you through the review process, from registering to be a reviewer through to leaving a review on a panel.
12.12.2018 Thank you! We have completed all the Version 1+ gene panels required to cover all the rare diseases in the 100,000 Genomes Project! #100KThankYous
This comprises of:
- 174 public gene panels
- Covering almost 50% of OMIM phenotypes
- 13,875 gene-disease curations
- 4423 individual genes
- 7346 high-level of evidence diagnostic-grade "Green" genes used for genome interpretation
- Contributions from 206 different reviewers worldwide
These panels are all publicaly available and queriable. They are dynamically updated by our Curation Team...we encourage experts within these disease areas to provide reviews or add new genes as new evidence arises!
This panel was formed from the merger of the following Version 1+ panels: Epileptic encephalopathy (Version 1.132)(code 67), Familial Focal Epilepsies(Version 1.9)(code 252), Familial Genetic Generalised Epilepsies (Version 1.23)(code 240) and Genetic Epilepsies with Febrile Seizures Plus (GEFS+) (Version 1.9)(code 160). Genes from the pre-version 1 Epilepsy Plus (Version 0.14)(code 161) panel were also included. All these former panels have now been retired.
We would like to thank all reviewers and curators for their contribution to this panel, with especial thanks going to Zornitza Stark (Victorian Clinical Genetics Services) for sharing her gene lists and expertise.
This gene panel is designed to cover several limb disorders, where limb disorder is the primary or secondary feature, including Brachydactyly, Polydactyly, Syndactyly, Ulnar ray abnormalities and Radial ray abnormalities. Thank you to our external reviewers and Genomics England Clinical Fellows for helping to get this panel completed.
Thank you to Ioannis Sarantitis (EUROPAC) for providing an initial gene list and expert review of the panel.
The Green genes on this panel will be utilised in the Genomics England Rare Disease analysis pipeline for genome interpretation. We would like to send a big thank you to Bill Griffiths (Cambridge University Hospitals) for providing gene lists and expert review to aid the curation of this panel.
20.11.2018 Short QT syndromes for rare diseases is now Version 1.
This version of the Short QT panel has been formed from the expert list provided by Jules Hancox (University of Bristol), together with re-evaluation with respect to Short QT, of the genes from the Long QT syndromes panel (Version 1.5)(https://panelapp.genomicsengland.co.uk/panels/76/) and from the Brugada syndrome panel (Version 1.7)(https://panelapp.genomicsengland.co.uk/panels/13/). Thank you very much to Jules Hancox for his considerable contribution to this panel.
This panel has been promoted to V1 after extensive external expert review, internal review and curation, together with Genomics England clinical input. The Non-syndromic familial congenital anorectal malformations panel will be used to aid genome interpretation for participants recruited under this disease.Thank you to all our external reviewers and Genomics England Clinical Fellows.
11.10.2018 New PanelApp Handbook
We have updated the PanelApp Handbook to guide you through all our new features, including:
-
The new API
-
Copy Number Variants (CNVs) from the curated ClinGen Dosage Sensitivity Map curated regions
-
Short Tandem Repeats (STRs)
-
Panel types and cancer susceptibility germline gene panels
-
Super panels
-
Panel activity
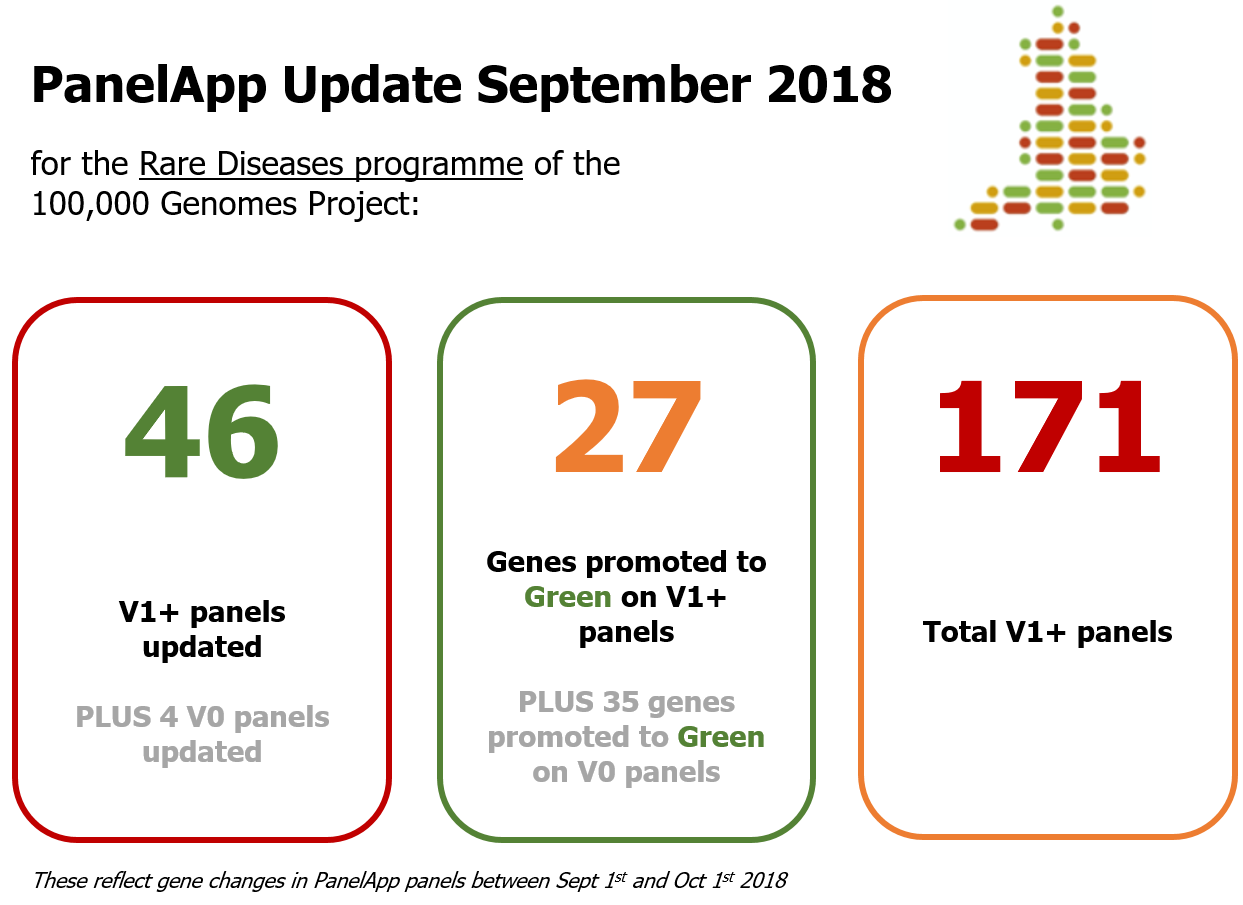
02.10.2018 . We have been busy updating panels in PanelApp last month. Here's a summary of panel updates for September 2018:
PanelApp Upgrade Release v2.3.0
New Features:
Copy Number Variants
Support for CNVs has been added to PanelApp, allowing these to be integrated into our genome analysis pipeline for rare diseases. We have now added:
These originate from the curated ClinGen Dosage Sensitivity Map curated regions, available here.
Find CNVs in PanelApp on the Genes and Entities page by filtering for ISCA code or view on individual panels, for example the Primary Microcephaly - Microcephalic Dwarfism Spectrum gene panel.
Each CNV has the following features - see this example:
-
An ISCA code and a more verbose name that is more commonly recognised
-
Is denoted as either a loss or a gain.
-
Is tagged as a 'Region' in the panel - you can therefore filter the panel to view only regions.
Panel Types
Each panel now has a 'Panel Type' which appears on the panel page. This helps distinguish which gene panels belong to which projects.
Current panels fall under:
The webservices a query for all panels (https://panelapp.genomicsengland.co.uk/WebServices/list_panels/)
will now by default return a list of those panels that have the Rare Disease 100K panel type.
Super panels
Support for creating a ‘Super panel’ has been added to PanelApp for Genomics England Curators. This allows us to create a parent panel made up of a group of child panels. Whenever a child panel is updated, the Super panel is also updated (and the version number on both the child and Super panel is increased). In cases where there is the same gene in multiple child panels, the Super panel will display two rows with links to separate entities in their respective panels. For WebServices, when querying the Super panel, all data from child panels will be returned.
Super panels will be added in the next few months so watch this space!
Activity
This release of PanelApp includes improved recording and searching of changes to panels:
-
View the activity on individual panel pages by clicking on the ‘Panel Activity’ button found under the panel description box. An example is given here for the intellectual disability panel; this shows the latest activity on this panel, and can be further filtered by gene/genomic region, person, activity type by typing in the filter box e.g. ‘new gene’.
-
You can now filter the Activity page: click on the filter icon to select the panel or date you would like to view (version and gene/genomic entity to come soon). Filter this further by typing in the activity type, gene, or person in the filter box.
5.09.2018 PanelApp will be upgraded from v2.20 to v2.30 from 10am - 12pm (BST) this Friday, September 7th. Panelapp will not be accessible during this time.
New features will include:
-
Region functionality: Support for regions (such as copy number variants). Currently, gain and loss are the two types supported.
-
Panel Types: Each panel will now have a Panel Type. This will help distinguish which panels belong to which projects. You will see a Rare Disease 100K and Cancer Germline 100K panel type for the Genomics England Rare Disease 100,000 Genomes Project and the Genomics England Cancer 100,000 Genomes Project.
-
Super panels: Support for Super panels has been included. A Super panel is made up of a group of child panels. Whenever a child panel is updated, the Super panel is also updated.
-
All activity on panels recorded: Improved recording and searching of changes to panels via filtering, which can be viewed in the Activity tab, including when curators or external reviewers add genes, phenotypes, mode of inheritance, etc
3.09.2018 Neonatal cholestasis for rare disease is now Version 1.
This panel has been promoted to V1 after extensive external expert review, internal review and curation, together with Genomics England clinical input. Neonatal cholestasis panel will be used to aid genome interpretation for participants recruited under this disease.Thank you to all our external reviewers and Genomics England Clinical Fellows.
PanelApp Release v2.2.0: We have expanded the scope of PanelApp to allow curation and review of Short Tandem Repeats (STRs) associated with disease. This guide provides an overview to STRs and the changes to PanelApp
- The 'Genes' page is now 'Entities'
- Reviewers can review and add STRs to a panel
- Downloads of panels include both genes and STRs
This panel has now been promoted to Version 1 after external expert review, internal curation and clinical input and will be used to aid genome interpretation for participants recruited under this disease.Thank you to our external reviewers, in particular Sophie Hambleton for her extensive reviews and assistance with this panel.
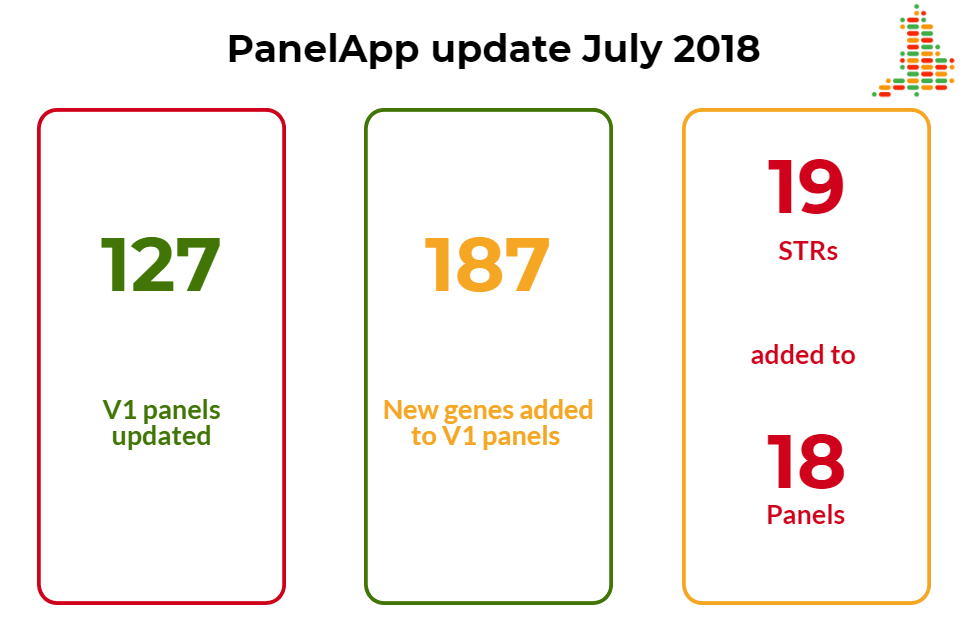 A review of the year up to July 2018
A review of the year up to July 2018
09.04.2018 Biocuration 2018
Our Scientific Curator Eleanor Williams is attending the International Biocuration Conference in Shanghai, China and will be presenting her Career talk on Wednesday. Look out for @PanelAppTeam tweets regarding interesting aspects of the conference.
27.03.2018 Our Scientific Curator Olivia Niblock presented a poster at the Genomics of Rare Disease conference
The poster outlined the PanelApp curation process and review tool.
This panel has now been promoted to Version 1 after external expert review, internal curation and clinical input and will be used to aid genome interpretation for participants recruited under this disease. Thank you to our external reviewers Georgios Korres, Jose Antonio Lopez-Escamez and Maria Bitner-Glindzicz for evaluating this gene panel!
23.03.2018 New diagnostic-grade gene panel available
The Idiopathic ventricular fibrillation gene panel has been promoted to Version 1 after external expert review, internal curation and clinical input. For patients recruited under this disease to the 100,000 Genomics Project, the following additional gene panels will also be applied for genome analysis:
12.03.2018 ID Gene Panel Update Phase III
Today the Intellectual disability gene panel was promoted to Version 2.0 to reflect major updates that have occured since November 2017:
-
Reviews for 836 genes have been added from the Genomics England Curators and the Clinical Team.
-
127 new Green genes have been added to the panel for genome interpretation (from 754 to 881 Green genes).
-
The gene total has increased from 1879 to 1927.
07.03.2018 Panelapp was upgraded to v2.1.2.
All panels now have a new field called “status” via the WebServices get_panel and list_panels endpoints. Now we have an enumeration which includes the status of the panel as shown below:
-
public (visible to the public)
-
retired (visible in the webservices, not visible via website); these panels are those that have previously been used for genome interpretation, however have now been retired as they are now merged into another public gene panel. These panels will no longer be used independently for interpretation, however are accessible for reference.
In addition, there has been a fix to genes missing an Ensembl gene ID.
This gene panel was created for application to rare disease participants recruited with multiple tumours or young adult onset cancer. The green genes on this panel will be used to aid genome interpretation for these participants. Thank you to Clare Turnbull and Helen Brittain for creating and evaluating this gene panel.
28.02.2018 PanelApp now on Twitter
You can now follow us on twitter @PanelAppTeam #PanelApp to get the latest news about our virtual diagnostic gene panels and keep up to date with the project, find out when new panels are added and for which panels we are recruiting reviewers. In addition we will provide information about improvements to the PanelApp tool and any planned downtime.
Thank you to the reviewers for making this possible.
20.01.2018 View and filter updates to PanelApp on the Activity page
23.01.2018 Congratulations to our new Curator Eleanor Williams who has been awarded this year's Biocuration Society Career Award!
We are happy to announce that Eleanor has just joined the Genomics England PanelApp Curation Team, bringing a wealth of Biocuration experience. Read more about her career and her valuable work that has led to this award.
A talk entitled "The 100,000 Genomes Project Rare Disease Programme: Achievements and Future Plans"
was presented by Dr Emma Baple, Clinical Lead for Rare Disease Validation and Feedback at Genomics England. Her talk included how PanelApp is used for genome analysis and diagnosis of rare diseases. On PanelApp we now have over 800 registered reviewers from around the world - including those from Arab countries.
05.01.2018 Intellectual disability gene panel update Phase II
The following major updates were made to the ID gene panel:
- Reviews for 290 genes by Genomics England Curators and Clinical Team were added.
- Gene status for these genes was updated according to evidence level, resulting in 41 new Green genes (858 in total).
- The number of total genes on the panel was increased from 1895 to 1911.
- Updated version: 1.625 (previous version before updates began was 1.561).
03.01.2018 EDS Society news post regarding PanelApp
Read this blog post regarding our EDS gene panel, reviewed by members of the EDS Society.
PanelApp holiday closure Friday 22nd December 2017 to 2nd January 2018
Dear PanelApp Users - please note that Genomics England offices will be closed from Friday 22nd December 2017 until 2nd January 2018, and so there may be a delay in response to registration requests or emails to [email protected] - PanelApp will still be available during this time for you to review, download or query panels. We would like to thank all our Reviewers and wish a happy holiday to all our Users
15.12.2017 Primary Membranoproliferative Glomerulonephritis promoted
The PMG panel was promoted to Version 1 - the green genes on this panel can now be used for genome interpretation - thank you to Dr Daniel Gale, University College London, Arianna Tucci (Genomics England Clinical Team) and Louise Daugherty (Genomics England Curation Team) who curated and reviewed the evidence for the genes on the panel.
29.11.2017 Major update to the Intellectual disability gene panel
Today we made a major update to the ID panel, adding extensive reviews of 383 genes by Curators and Clinical fellows at Genomics England who have been investigating the evidence behind these genes in the last few months. The gene rating for these genes was updated according to evidence level, resulting in 66 new Green genes (817 in total) which will be used for genome interpretation. Publications, comments and decisions made based on the evidence can be viewed under the 'Reviews' tab of a gene. The number of total genes on the panel was increased from 1879 to 1895.
17.11.2017 New gene panels
We have added the following panels to PanelApp - if you have expertise in these areas please help us develop these panels by adding genes and reviews:
06.11.2017 We launch a new and improved release of PanelApp!
What's new?
A simpler URL https://panelapp.genomicsengland.co.uk
Direct URL links to panels or genes are available, even if you are not logged in
Straightforward links to genes e.g. https://panelapp.genomicsengland.co.uk/panels/101/CHD7/
Shorter panel codes e.g. https://panelapp.genomicsengland.co.uk/panels/101/
Both Genome build GRCh38 and GRCh37 are supported
This includes updates to some HGNC-approved gene symbols
New webservice queries are available; you can specify assembly GET parameters with either GRch37 (default) or GRch38 as a value. EnsemblIds will be returned for the specified assembly version: GRch37 version 82 or GRch38 version 90 if they exists in the database. For example https://panelapp.genomicsengland.co.uk/WebServices/search_genes/AKT2/?panel_name=Regional%20overgrowth%20disorders&assembly=GRch38
Improved page loading and greatly improved response times
Improvements to the registration process
02.11.2017 A summary of updates to Version 1 panels in October
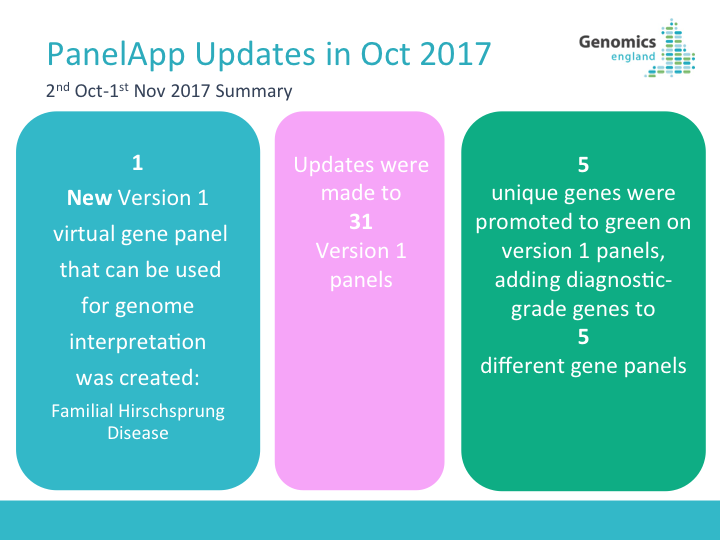
30.10.2017: PanelApp Update: 167 diagnostic-grade (Version 1+) panels

20.10.2017 A summary of updates to panels in September
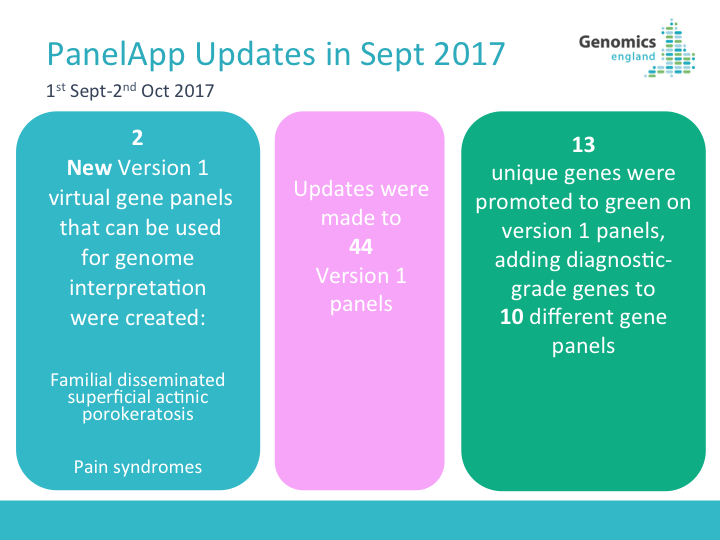
19.09.2017: Diagnostic-grade Pain syndromes gene panel released
A combined panel for Mendelian disorders of pain perception, including insensitivity to pain or increased pain perception has been promoted to Version 1; the green genes on this panel have a high level of evidence and can be used in genome interpretation.
29.08.2017: PanelApp Update: 165 diagnostic-grade (Version 1+) panels

31.07.2017: PanelApp Update
The Ehlers-Danlos syndromes panel is now Version 1 and ready for patient interpretation!
The number of 'Amber/Watchlist' genes on our Version 1 panels has also taken a jump thanks to a large update of the Intellectual disability panel. Here's a summary of our current Version 1 gene panel statistics:

27.07.2017: Hirschsprung's disease in the 100,000 Genomes Project.
Our Curator Rebecca Foulger is attending the 3rd Hirschsprung's Disease Conference at Alder Hey Children's hospital, Liverpool, on Friday 28th July to present the ongoing Hirschsprung's gene panel. You can view and review the Familial Hirschsprung Disease panel here. The conference is organised by the charity CHAMPS appeal.
26.07.2017: 19 gene panels launched for reporting pertinent findings in cancer germline genomes
We have released genes panels for pertinent findings in cancer germline genomes, for patients recruited under different tumour types. A big thank you to Clare Turnbull (Clinical Lead for Cancer Genetics and Genomics for 100,000 Genomes Project, Genomics England) for collating these genes lists together, and seeking input from the clinical cancer community.
View and review them here:
20.07.2017: Two skin panels promoted to Version 1
The following two dermatological disorder panels have been promoted to Version 1, which will enable the green (high-level of evidence) genes to be used for genome interpretation. Thank you to Professor John McGrath at King's College London for expert review.
04.07.2017: Latest update of Rare Disease Eligibility Criteria V1.7.2
28.06.2017: PanelApp Presentation
Ellen McDonagh, Head of curation at Genomics England, is currently attending the Curating the Clinical Genome conference in Washington. Ellie is giving a talk on PanelApp and 'The Impact of Community Curation of Gene- Disease Relationships for Clinical Genome Analysis'.
26.06.2017: PanelApp Update:

15.06.2017: Expert review needed for skin disorder gene panels:
We are currently seeking expert review for the following dermatological disorder panels:
If you have expertise in this area and are able to provide input into these two panels, or have any questions about the panels, please contact [email protected].
06.06.2017: Version 1+ gene panels are ready for patient interpretation!

31.05.17: Promotion of 6 gene panels to version 1 - now available for genome interpretation
Our curators have been working hard to promote 6 more gene panels to version 1, enabling the green high-level of evidence genes to be used for genome interpretation - thank you to all external reviewers who helped with this achievement:
23.05.17: The latest panel requiring external review is Inherited non-medullary thyroid cancer. Please contact us if you think that you could help with this.
23.05.17: Diagnostic-grade panel for Radial dysplasia
Our latest gene panel to be promoted to Version 1 is Radial dysplasia. This panel contains 58 genes that have been sourced and reviewed, including 47 "green" (diagnostic-grade) genes which can be used for the analysis of patient genomes.
15.05.17: Learn more about PanelApp in this blog by James Hadfield
James Hadfield, Head of Genomics at CRUK Cambridge Institute, has written a short Enseqlopedia blog about PanelApp. Get in touch with us if you have any questions!
Panels requiring external expert review (8th May 2017):
14.05.17: A unified GI tract panel
Ellen, Sarah and our team of reviewers has been working on a combined panel for GI track tumour syndromes, combining three previous panels: Familial colon cancer, Multiple bowel polyps, and Peutz-Jeghers syndrome. The panel is now Version 1, and the diagnostic-grade (green) genes can be used for genome analysis of patients in the 100,000 Genomes Project.
10.05.17: 153 Version 1+ panels!
We now have revised diagnostic-grade green gene lists that can be used for genome analysis for the following diseases:
08.05.17: 151 Version 1+ panels!
Severe hypertriglyceridaemia is our latest gene panel to be promoted to Version 1. That takes our total Version 1+ gene panels to 151!
150 Version 1+ panels!
After our 6th Gene Panel Curation Day last Thursday, we now have 150 Version 1+ panels ready for the analysis of patient genomes, including:
Developmental Glaucoma,
Familial pulmonary fibrosis and Epidermolysis bullosa
Thank you to all our reviewers, curators and clinicians!
02.05.17 PanelApp update
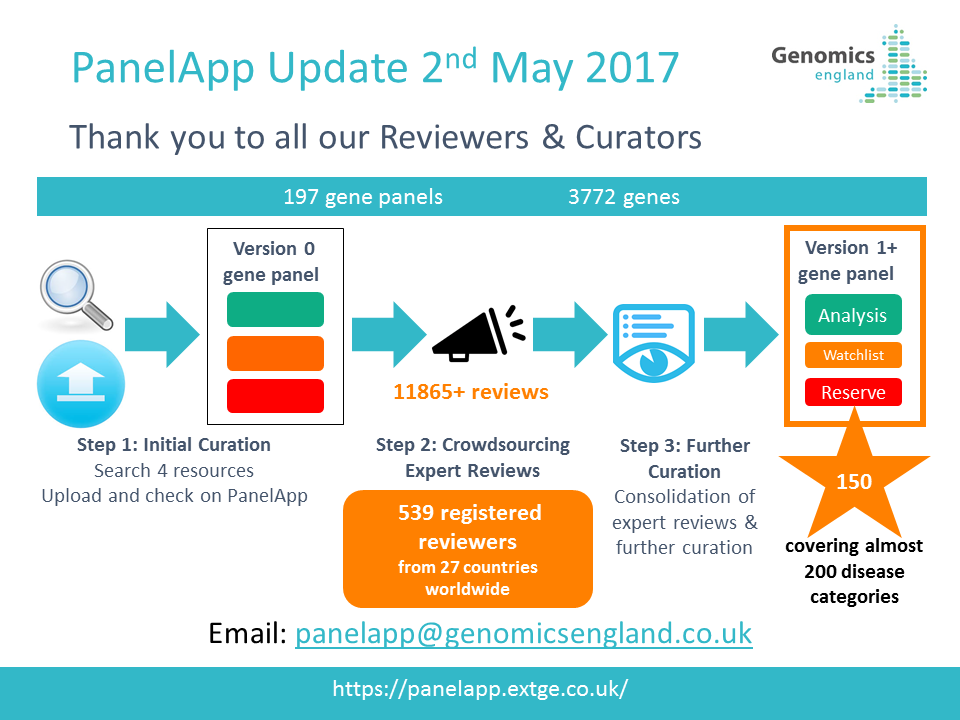
Our 6th Gene Panel Curation Day
On Thursday 27th April 2017, our Curation and Clinical team will get together at the Wellcome Genome Campus, Hinxton for our 6th Gene Panel Curation day! We'll work together to curate and finalise Version 1 gene panels ready for the analysis of patient genomes. We'll keep you updated on our progress!
25.04.17 PanelApp update
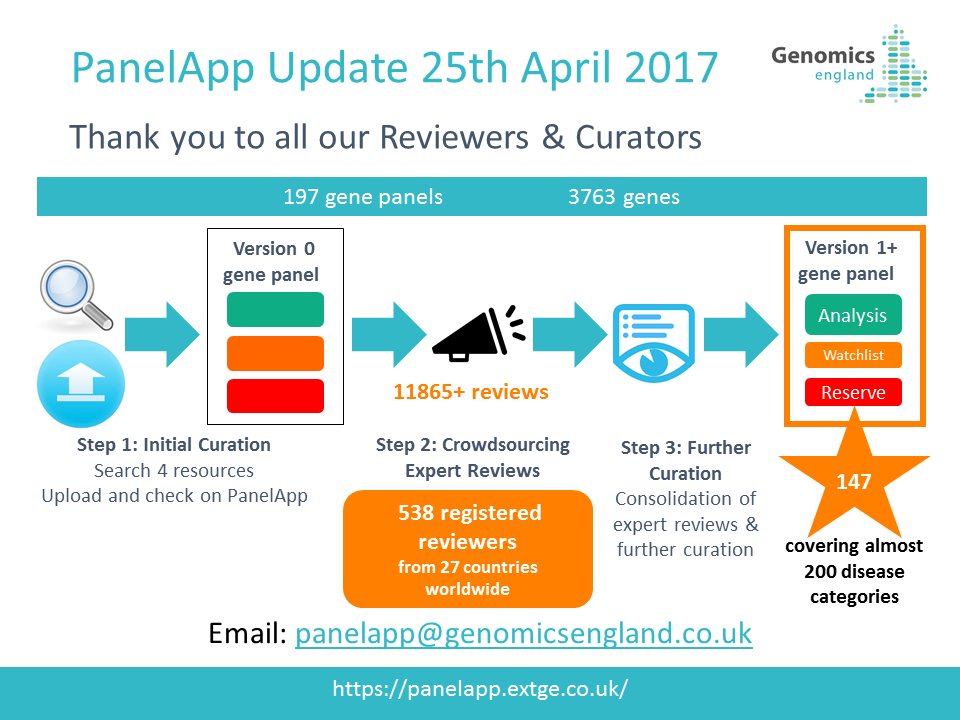
18.04.17 PanelApp update
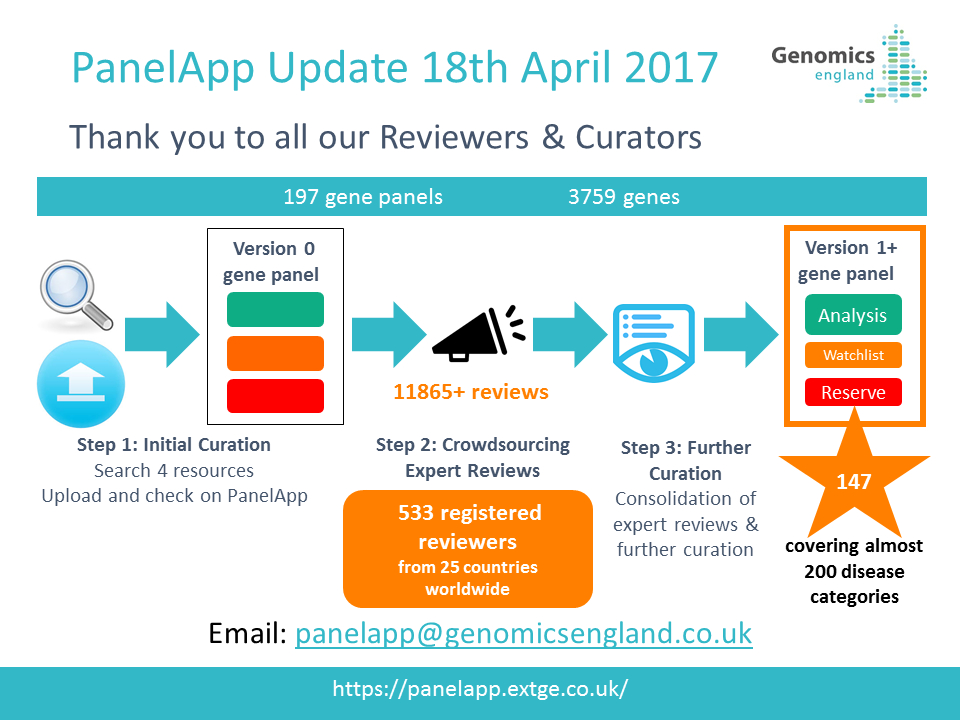
12.04.17 PanelApp webservices (API) are available to query using the examples available on the PanelApp How To tab.
Some gene panels may have been applied for genome analysis and interpretation reports that are now retired and no longer live in webservices or on the PanelApp UI. To access a previously used panel version, use the panel name (rather than the code) and the version number to retrieve the correct panel.
An example is the panel for Bilateral microtia (which is retired on PanelApp because it has now been merged into the Deafness and congenital structural abnormalities panel but can be retrieved by webservices using the specific version number):
/crowdsourcing/WebServices/get_panel/Bilateral%20microtia/?version=1.4
10.04.17 PanelApp update
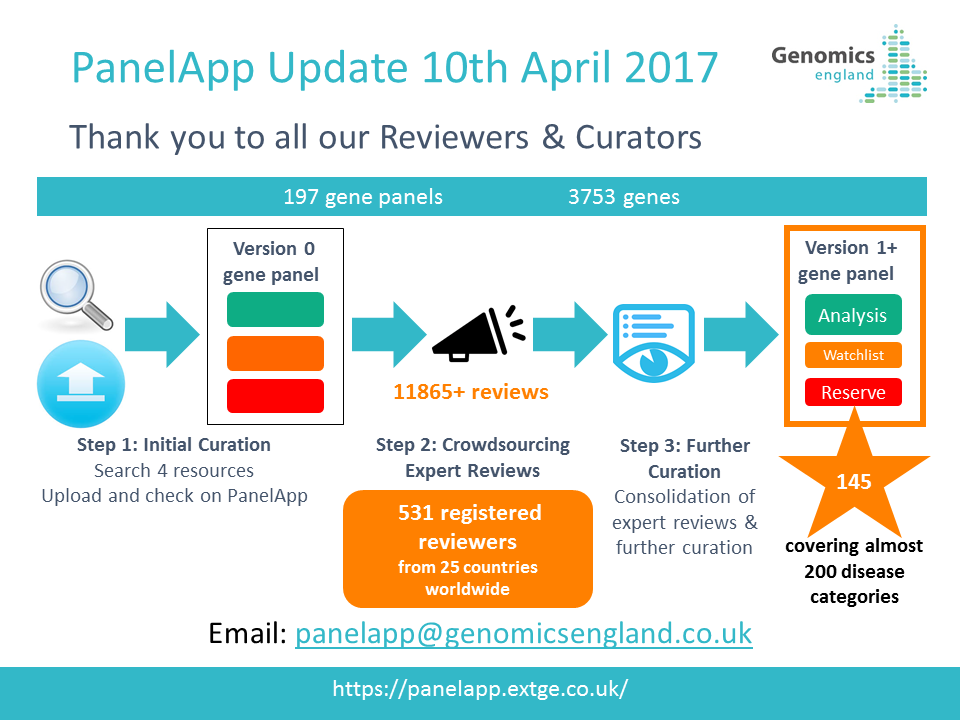
06.04.17 Rebecca is presenting the poster "PanelApp: A Key Resource for the Rare Disease Community" at the 'Genomics of Rare Disease' conference, Wellcome Genome Campus, Hinxton today (Poster 14) #GRD17
03.04.17 PanelApp update
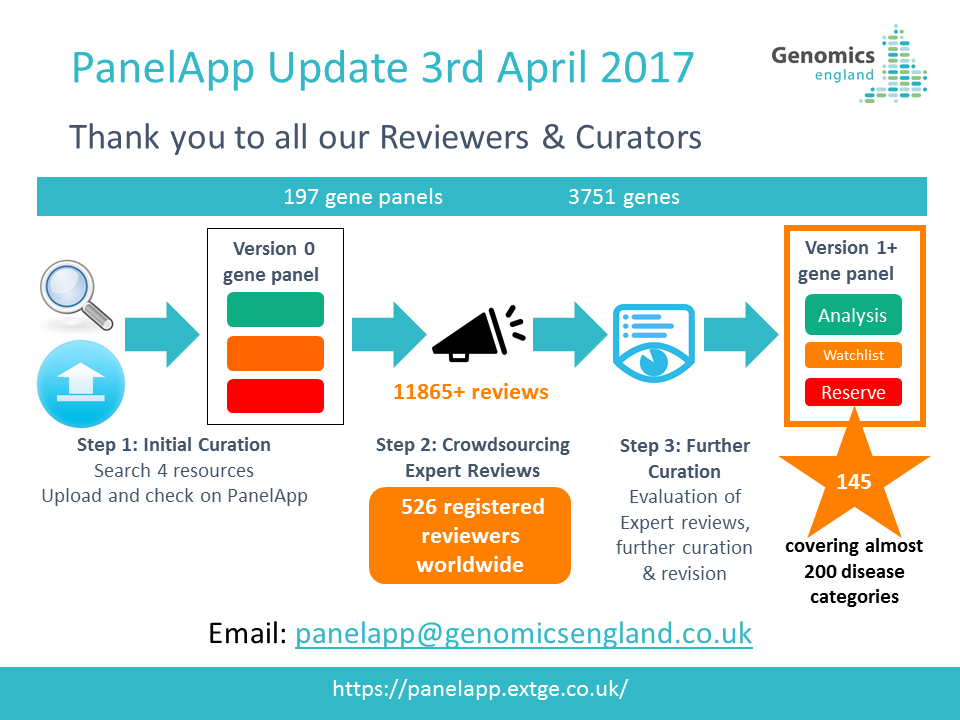
29.03.17 "The Impact of Community Curation on Rare Disease Diagnosis" was presented at the 10th International Biocuration Conference, Stanford University by Ellen McDonagh
The presentation included an overview of PanelApp and the curation-crowdsourcing process for creation of rare disease panels.
21.03.17 PanelApp update
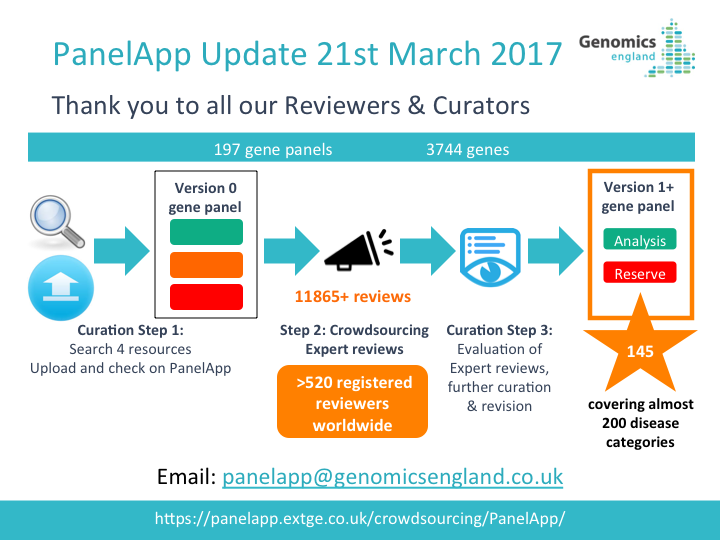
13.03.17 PanelApp update
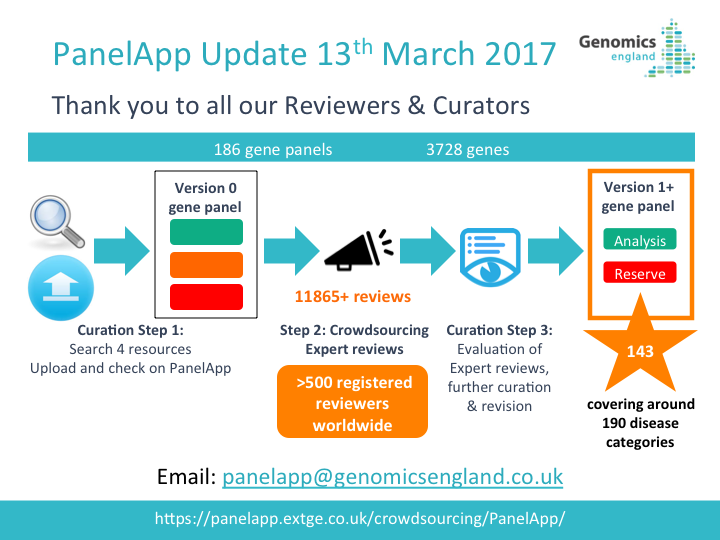
06.03.17 PanelApp update

23.02.17 PanelApp update
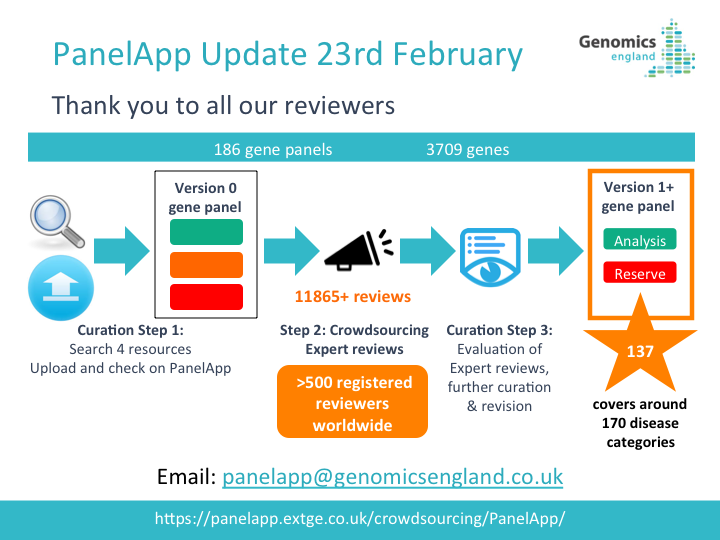
22.02.17 Our 5th Gene Panel Curation Day
Our Curators, Clinical Fellows and Clinical Geneticists got together for the day to discuss genes with difficult scenerios, curate and finalise gene panels: we now have 137 reviewed and revised gene panels! (These are version 1+).
A big thank you to all reviewers who have contributed to these panels, and to everyone involved in the day:
Sarah Leigh, Rebecca Foulger, Olivia Niblock, Louise Daugherty, Alice Gardham, Richard Scott, Arianna Tucci, Helen Brittain, Chris Campbell, Ellen Thomas, Damian Smedley, Ellie McDonagh. Thank you also to Antonio Rueda-Martin for essential technical assistance.
The following 7 panels were promoted to version 1 and the green 'diagnostic-grade' genes can now be used for analysis of genomes:
17.02.17 PanelApp Update
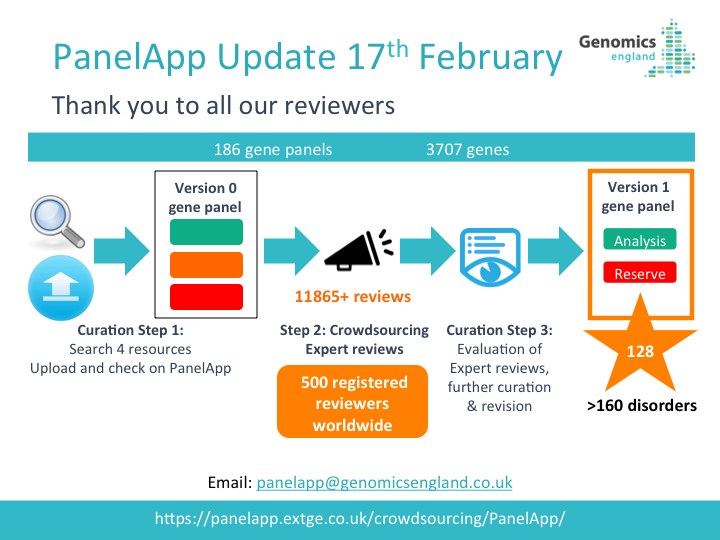
Please see the 41 panels that require expert review:
08.02.17 BRIDGE consortium Tier 1 genes from NIHR BioResource - Rare Diseases Study (NIHRBR-RD) added to PanelApp
We have added the Tier 1 gene list from Inherited bleeding disorders project
(BPD). This panel is designed to cover rare inherited bleeding disorders, inherited platelet and thrombotic disorders.
The panel contains green reviews corresponding to BRIDGE consortium Tier 1, as evaluated by the following experts from the BRIDGE consortium NIHRBR-RD contributed to this panel:
-
Prof Willem Ouwehand, Director NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
-
Dr Keith Gomez, Royal Free Hospital, London
-
Prof Kathleen Freson, Centre for Molecular and Vascular Biology, Leuven, Belgium
-
Prof Michael Laffan, Imperial College, London
-
Dr Andrew Mumford, University of Bristol
-
Dr Ernest Turo, NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
-
Karyn Megy, NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
-
Louise Daugherty, NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
Thank you to Karyn Megy, WGS Clinical Feedback Lead for the NIHR BioResource - Rare Diseases Study (NIHRBR-RD) for providing this list, and to the experts who contributed to the review.
We plan to add further BRIDGE consortium gene lists to PanelApp: watch this space!
07.02.17 BRIDGE consortium Tier 1 genes from NIHR BioResource - Rare Diseases Study (NIHRBR-RD) added to PanelApp
We have added the Tier 1 gene list from Specialist Pathology: Evaluating Exomes in Diagnostics project (SPEED_NEURO) which covers epilepsies, movement and microcephaly disorders.
The panel contains green reviews corresponding to BRIDGE consortium Tier 1, as evaluated by the following experts from the BRIDGE consortium NIHRBR-RD contributed to this panel:
-
Professor F. Lucy Raymond, Cambridge Institute for Medical Research, University of Cambridge
-
Manju Kurian, Paediatric neurologist, Great Ormond Street Hosptial
-
Keren Carss, NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
-
Alba Sanchis-Juan, NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
-
Marie Erwood NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
-
Louise Daugherty, NIHR BioResource - Rare Diseases, Cambridge University Hospitals NHS Foundation Trust
Thank you to Karyn Megy, WGS Clinical Feedback Lead for the NIHR BioResource - Rare Diseases Study (NIHRBR-RD) for providing this list, and to the experts who contributed to the review.
We plan to add further BRIDGE consortium gene lists to PanelApp: watch this space!
06.02.17 We now have 500 registered reviewers!
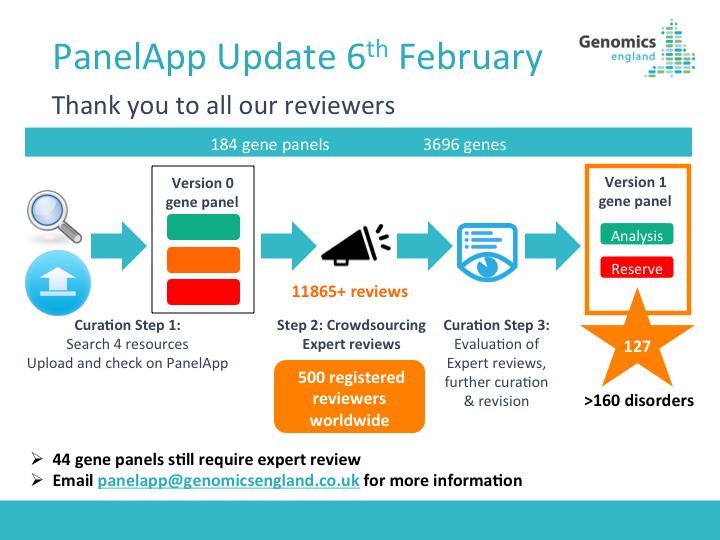
27.01.17 PanelApp Update
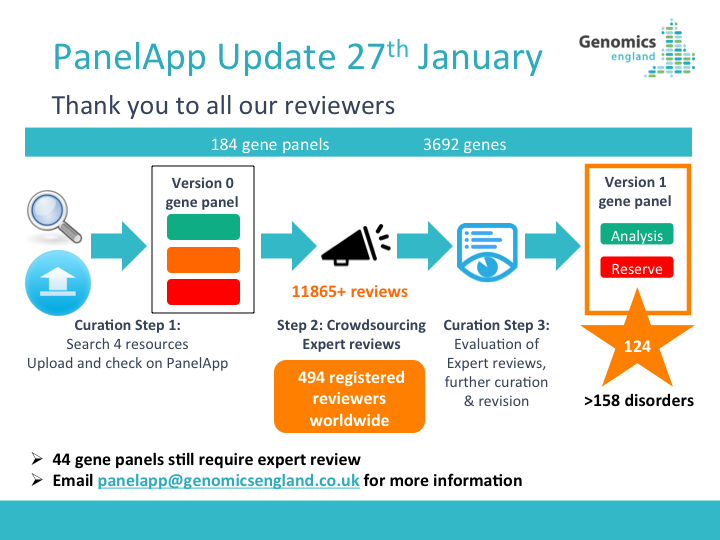
This panel combines genes for Classical Ehlers-Danlos Syndrome, Kyphoscoliotic Ehlers-Danlos syndrome, Ehlers-Danlos Syndrome (unusual phenotypes e.g. absent pain sense) and Ehlers-Danlos syndrome type 3 together. Reviews from these individual panels have been transferred over. Please help us review the genes on this panel to create a diagnostic-grade list of 'green' genes.
23.01.17 Three new disorders have been added to the rare disease list for the 100,000 Genome Project
We have created placeholder gene panels for:
Please click on the panel to add genes and provide your review.
17.01.17 Our first version 2 panel is realised
The Arthrogryposis gene panel was revised to include an additional 101 green genes based on information from diagnostic labs and further curation. As this was such a major change from 10 green genes, the panel was promoted to the next major version 2.0.
10.01.17 PanelApp Update
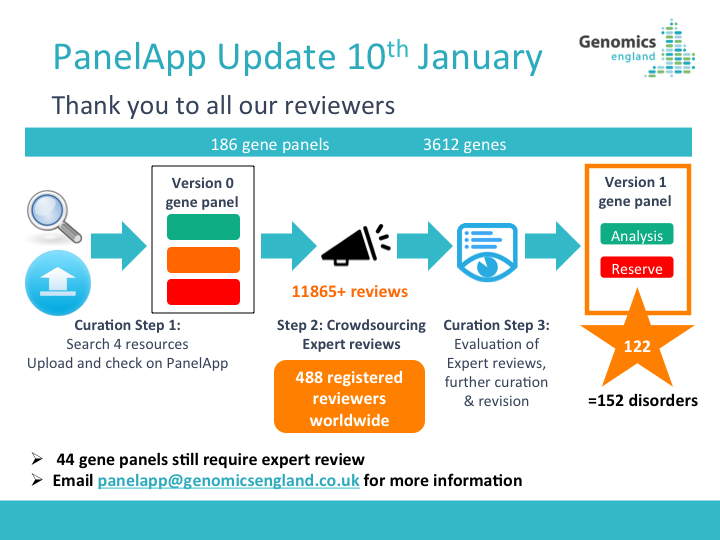
44 gene panels still require expert review - if you have expertise in one of these disease areas, please register or sign in to review the genes on the panel to help us establish a diagnostic-grade green list:
03.01.17 Happy New Year to our PanelApp users!
We start the year with 121 version 1 panels covering 147 disorders with a diagnostic green list of genes and a total of 3605 genes. The 65 panels that still require expert review and internal update can be found here:
/panels/
We would like to thank all our expert reviewers who contributed to PanelApp in 2016, and welcome those providing reviews this year.
19.12.16 The 4th PanelApp Gene Panel Curation Day
This is when the PanelApp Curators get together in a room with the Clinical Geneticists at Genomics England and revise gene panels according to expert reviews and further curation according to our internal rule set. By getting together in the same room, we can discuss any complicated gene scenerios or evidence issues. Once a panel has been revised, we promote it to version 1; the green genes on this panel will then be used by our Bioinformaticians to help analyse genomes and narrow down potentially diagnostic variants for the 100,000 Genomes Project.
We now have 118 version 1 panels, having promoted 9 gene panels at the Curation Day:
And revised the Malformations of cortical development gene panel to add further genes after an additional expert review.
05.12.16 PanelApp update
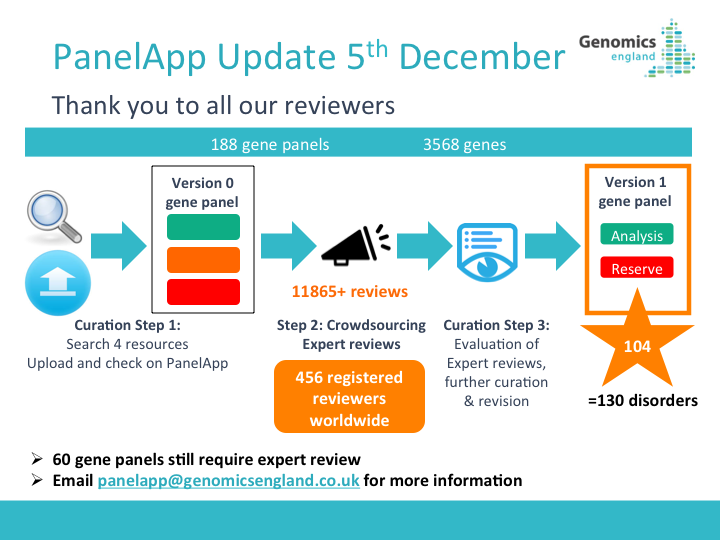
18.11.16 We now have 100 version 1 gene panels! The green genes from these panels are being used to help prioritise variants within patient genomes recruited for the 100,000 Genomes Project.
We also now have 456 reviewers from institutions worldwide - a big thank you to those who have signed up and are reviewing our gene panels!
14.11.16 Our curation team is expanding!
Rebecca Foulger joined Genomics England as a Scientific Curator at the start of October. Rebecca has worked on ontology and database projects since her PhD, including FlyBase, UniProt and the Gene Ontology (GO) project, and she joins us from her Parkinson's disease GO annotation project at UCL.
26.10.16 We now have 95 version 1 gene panels! The green genes from these panels are being used to help prioritise variants within patient genomes recruited for the 100,000 Genomes Project.
The following panels were recently promoted:
06.10.16 We now have 90 version 1 gene panels, of which the green genes are being used to analyse genomes of participants recruited in the 100,000 Genomes Project
Thank you to all the expert reviewers and curators who contributed to these panels. They can be viewed on the panel page by filtering with 'version 1'.
27.09.16 PanelApp was presented both in the UK and in California today
-
PanelApp and the gene panel curation process was presented at the UKGTN Clinical & Scientific Group Meeting in London by Sarah Leigh.
-
PanelApp featured in an overview of the 100,000 Genomes Project presented at Illumina in Santa Clara, California by Ellen McDonagh.
26.09.16 PanelApp was presented in a talk at Stanford University, California, by Ellen McDonagh, in a presentation about the 100,000 Genomes Project.
22.09.16 An overview of the 100,000 Genomes Project was presented by Ellen McDonagh at Genentech, South San Francisco, California, with a focus on the role of the Bioinformatics team and how gene panels on PanelApp are used in the analysis of rare disease genomes for the project.
20.09.16 Ellen McDonagh presented the role of PanelApp as part of a panel discussion about crowdsourcing resources in genomics at the Festival of Genomics, San Diego, California.
16.09.16 PanelApp and an overview of the 100,000 Genomes project was presented by Ellen McDonagh to the Illumina Curation Team, Santa Clara, California.
13.09.16 We now have 87 Version 1 panels!
23.08.16 Updates to Version 1+ panels
4 green genes were added to the Hereditary Ataxia panel Version 1.7:
Changes to the Diabetes with additional phenotypes suggestive of a monogenic aetiology Version 1.1 were made due to feedback from an Expert Reviewer:
- AGPAT2 added and promoted to green (biallelic)
- BSCL2 added and promoted to green (biallelic)
- LRBA added and promoted to green (biallelic)
- PIK3R1 added and promoted to green (monoallelic)
- PLIN1 added and promoted to green (monoallelic)
- POLD1 added and promoted to green (monoallelic)
- SLC29A3 added and promoted to green (biallelic)
- TRMT10A added and promoted to green (biallelic)
- DCAF17 added as red (biallelic)
- IL2RA added as red (biallelic)
- PCBD1 added as red (biallelic)
- STAT1 added as red (monoallelic)
- IER3IP1 promoted from red to green (biallelic)
22.08.16 Happy Birthday PanelApp!
PanelApp was launched publically a year ago, on 21st August 2015. We now have 151 gene panels (and a placeholder for another 38 disorders), of which 56% have been reviewed and revised to Version 1, providing virtual gene panels for 104 disorders within the 100,000 Genomes Project. There are now being used to help analyse participant genomes. We would like to thank all Reviewers who have contributed their expertise and their time to PanelApp, and helped make this possible.
A big thank you goes to Antonio Rueda-Martin who created PanelApp, developed it and has kept it running successfully. Thank you also to Paul Hayes who has improved usability and created additional features.
Thank you also to all our Curators who have created panels, evaluated reviews and curated further information to revise and improved the panels.
15.08.16 We have 83 Version 1 panels, covering 102 disorders.
These panels have been reviewed and revised internally according to expert review and additional evidence. A big thank you to all reviewers who have contributed to these panels. To view these, go to the full gene panel list and filter for "version 1".
11.08.16 An update about PanelApp will be presented at the UKGTN meeting today, by Ellen McDonagh.
10.08.16 We now have an Activity page, displaying changes made to panels and when reviews are made.
20.07.16 New features have been added to PanelApp, and improvements to usability have been made!
We show some of these features below...
- You can now view, search and filter all genes by clicking on the Genes Tab found at the top of the page:
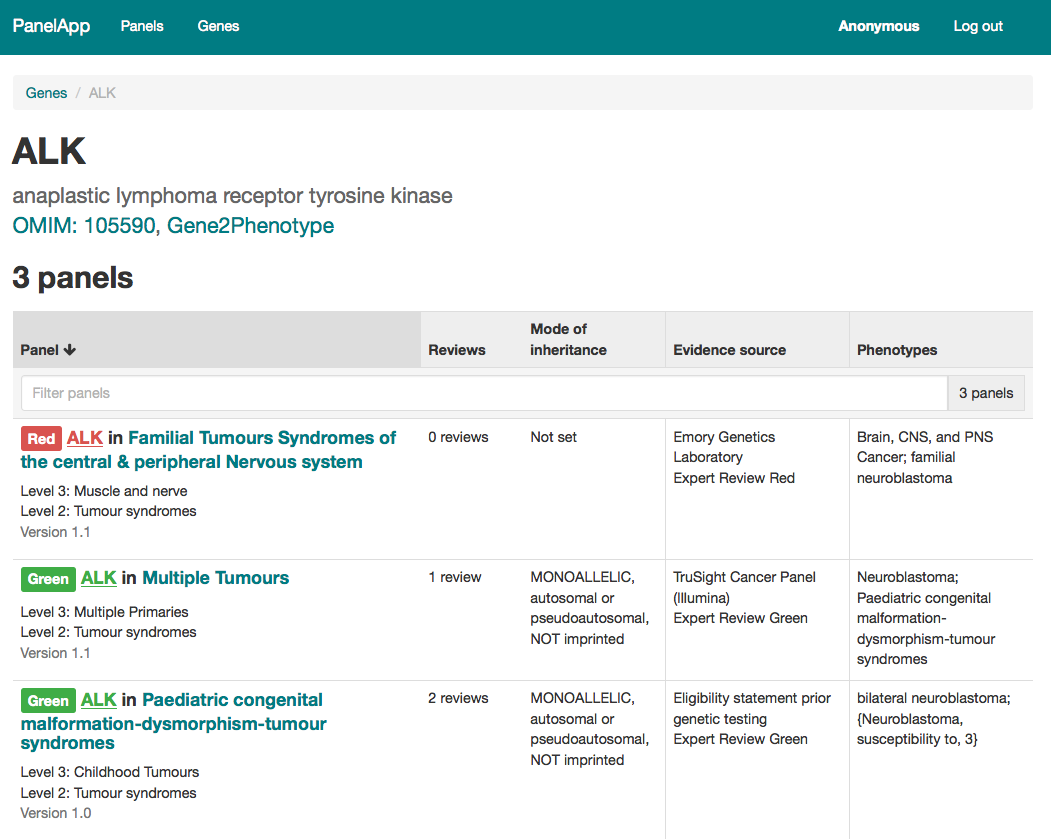
- You can add a review at the same time as adding a new gene to a panel:
- You can now compare two gene panels:
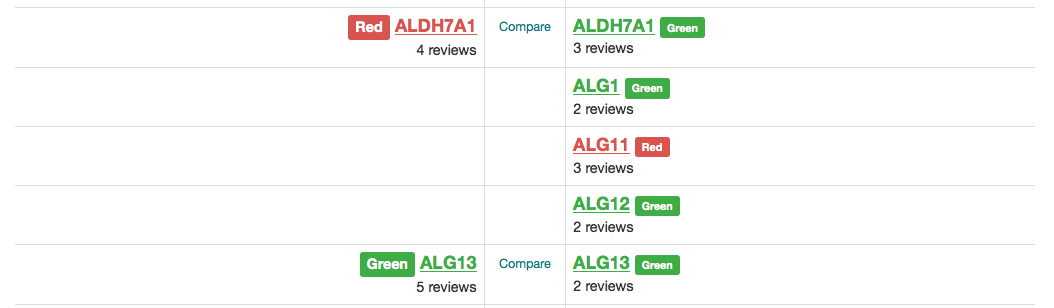
- There is a new look to the panel page:
22.06.16 PanelApp is presented at the Curating The Clinical Genome conference 2016 by Ellen McDonagh.
06.06.16 We now have 60 Version 1 gene panels!
These panels have been reviewed and revised internally according to expert review and additional evidence. A big thank you to all reviewers who have contributed to these panels. To view these, go to the full gene panel list and filter for "version 1".
02.06.16 The UKGTN contacted diagnostic lab heads endorsing PanelApp, requesting that labs submitting new NGS gene panel tests confirm the status of genes on PanelApp when relevant diseases are available, and provide reasons for excluding green genes/including red genes. They also encouraged labs to review genes to aid the process.
23.05.16 PanelApp was presented at the Pint of Science Festival to an audience in the Crown Pub in Clerkenwell, London, by Ellen McDonagh.
05.05.16 PanelApp was presented by Dr Katherine Smith at The European School of Genetic Medicine and European Society of Human Genetics Course in Next Generation Sequencing, Bertinoro di Romagna (Italy), May 4-7, 2016.
26.04.16 PanelApp Update
- We have >300 registered reviewers from around the world.
There are 53 (34%) gene panels with no review...please add your expert review to genes on these panels:
There are 18 (12%) gene panels with a partial review - please add your expert review to these panels to help gain a consensus:
There are 35 revised gene panels (Version 1), covering 46 of the approved rare disorders (22% of the total number of gene panels). Thank you to all reviewers who have contributed to these panels:
-
Hypertrophic Cardiomyopathy
-
Bardet-Biedl Syndrome
-
Hypertrophic Cardiomyopathy
-
Dilated Cardiomyopathy and conduction defects
-
Bilateral microtia
-
Multiple endocrine tumours
-
Brugada syndrome
-
Stickler syndrome
-
Agranulocytosis
-
Arrhythmogenic Right Ventricular Cardiomyopathy
-
Epileptic encephalopathy
-
Familial breast cancer
-
Intellectual disability
-
All recognised syndromes and those with suggestive features, includes Mitochondrial disorders and Lactic acidosis
-
Beckwith-Wiedemann syndrome (BWS) and other congenital overgrowth disorders, includes Atypical Beckwith-Wiedemann syndrome, Classical Beckwith-Wiedemann syndrome, Simpson-Golabi-Behmel syndrome, Sotos syndrome, Weaver syndrome
-
RASopathies, includes Noonan syndrome plus other features, Noonan syndrome, LEOPARD syndrome, Legius syndrome, Costello syndrome, Cardio-facio-cutaneous syndrome
-
CAKUT
-
Chondrodysplasia punctata
-
Congenital hearing impairment, profound/severe
-
Craniosynostosis syndromes phenotypes
-
Erythropoietic protoporphyria, mild variant
-
Familial Focal Epilepsies
-
Familial haematuria
-
Familial Thoracic Aortic Aneurysm Disease
-
Hydroa vacciniforme
-
Hyperinsulinism
-
Left Ventricular Noncompaction Cardiomyopathy
-
Long QT syndrome
-
Mucopolysaccharideosis, Gaucher, Fabry
-
Multiple bowel polyps
-
Multiple Epiphyseal Dysplasia
-
Multiple Tumours
-
Neuro-endocrine Tumours- PCC and PGL
-
Paediatric congenital malformation-dysmorphism-tumour syndromes
-
Parathyroid Cancer
18.04.16 Additional webservices are available, allowing you to search PanelApp by gene:
-
Search for panels by gene:
/crowdsourcing/WebServices/search_genes/BTK/
-
Search for a gene in one panel:
/crowdsourcing/WebServices/search_genes/BTK/?panel_name=Intellectual%20disability&format=json
-
Search by a gene, and pull out information only when it is rated green:
/crowdsourcing/WebServices/search_genes/BTK/?&LevelOfConfidence=HighEvidence&format=json
-
Search by gene from a particular source (e.g. UKGTN):
/crowdsourcing/WebServices/search_genes/BTK/?&Evidences=UKGTN&format=json
-
For all genes in a panel:
/crowdsourcing/WebServices/search_genes/all/?panel_name=Intellectual%20disability&format=json
15.04.16 PanelApp is now one of the databases included on the BioSharing site.
Biosharing is a curated, searchable portal of inter-related data standards, databases, and policies in the life, environmental and biomedical sciences.
The methods behind curating the gene panels and gaining expert knowledge through crowdsourcing were presented.
08.04.16 PanelApp website is now available within the N3 network, and so issues accessing it within the NHS should now be resolved.
We apologise for any inconvenience this may have caused, and appreciate the feedback we have had from users.
05.04.16 The BRCA1 gene within the intellectual disability panel was demoted from green to red.
30.03.16 Update on changes to gene panels
A combined Beckwith-Wiedemann syndrome (BWS) and other congenital overgrowth disorders gene panel was created, combining the individual gene panels for the following disorders together:
- Classical Beckwith-Wiedemann syndrome
- Atypical Beckwith-Wiedemann syndrome
- Simpson-Golabi-Behmel syndrome
- Sotos syndrome
- Weaver syndrome
Expert review of these panels was used to create Version 1 of the Beckwith-Wiedemann syndrome (BWS) gene panel.
24.03.16 Update on changes to gene panels
Reviews from experts for the following gene panels have been internally evaluated. Using the reviews and other sources, the gene panels have been revised by Genomics England Curators and upgraded to Version 1.0. The green genes of these revised gene panels will be used for analysis of genomes for patients recruited under these disorders. We would like to thank all reviewers who have contributed to this effort - names and affiliations of those who have contributed are provided on each gene panel page.
Other changes to panels:
Gene panels for Cone Dysfunction Syndrome, Developmental macular and foveal dystrophy, Inherited macular dystrophy, Inherited optic neuropathies, Leber Congenital Amaurosis / Early-Onset Severe Retinal Dystrophy, Rod Dysfunction Syndrome
and Rod-cone dystrophy were combined into a Posterior segment abnormalities virtual gene panel.
14.03.16 Please be aware that PanelApp was experiencing technical issues and may have been unavailable to users.
We apologise for any inconvenience this may haved caused - the issue is now fixed. Please contact [email protected] if you experience any difficulties accessing panels, or cannot see reviews you have made.
29.02.16 Rare Disease Day
Contribute to rare disease genome analysis by reviewing panels on PanelApp if you have an expertise in a particular disease or gene.
23.02.16 PanelApp was included in a presentation by Dr Emma Baple at the NHS Genomics Medicine Centre (GMC) National Event.
For the 100,000 Genomes Project Rare Disease Program, Emma explained how variants from genome analysis are tiered using the green genes from a relevant gene panel in PanelApp, highlighting the importance of expert review of the panels for feeding back results to GMCs.
19.02.16 Gene panel news
Today there were announcements in the press regarding the publication of a comprehensive sequencing assay for inherited cardiac conditions (ICC).
The genes in their table of well-characterised, disease-causing genes (Table 1) are all currently rated green (diagnostic grade) within the relevant panels on PanelApp:
The full published ICC panel contains disease-causing, putatively pathogenic, research and phenocopy genes (supplemental Table S1). Genes on the full ICC panel were compared against genes within the relevant panels on PanelApp. Several additional genes were added to the following panels:
Arrhythmogenic Right Ventricular Cardiomyopathy,
Brugada syndrome,
Catecholaminergic Polymorphic Ventricular Tachycardia,
Dilated Cardiomyopathy,
Dilated Cardiomyopathy and conduction defects,
Familial Thoracic Aortic Aneurysm Disease,
Familial hypercholesterolaemia,
Hypertrophic Cardiomyopathy,
Long QT syndrome.
We are asking experts to review the genes on the panels to determine whether there is enough evidence for these genes to be diagnostic-grade (and should be rated green), or whether it is a research gene ( and should be rated red).
18.02.16: We now have 300 registered reviewers!
11.02.16 Update on changes to gene panels
Reviews from experts for the following gene panels have been internally evaluated. Using the reviews and other sources, the gene panels have been revised by Genomics England Curators and upgraded to Version 1.0. The green genes of these revised gene panels will be used for analysis of genomes for patients recruited under these disorders. We would like to thank all reviewers who have contributed to this effort - names and affiliations of those who have contributed are provided on each gene panel page.
Other changes to panels:
These were updated from our original videos, due to the new design of the user interface in December 2015 by Paul Hayes and Antonio Rueda. Thank you very much to Luke Webster (QMUL) and Lisa Dinh (Genomics England) for helping to create and publish these videos. The voice is Ellen McDonagh, Lead Scientific Curator at Genomics England.
The same videos are also available on YouTube:
29.01.16:
-
The eligibility statement for Familial Thoracic Aortic Aneurysm Disease was updated, and the TGFBR2 gene was added to the eligibility statement prior genetic testing genes.
-
A gene within the prior genetic testing for the Multiple bowel polyps panel was corrected from "MHS6" to "MSH6".
19.01.16: PanelApp webservices are now available to query using the following examples...
- Get a list of the panel names:
/crowdsourcing/WebServices/list_panels
- Get individual panels (last version of the panel by default):
/crowdsourcing/WebServices/get_panel/Epileptic%20encephalopathy/
- Select a specific version of a panel (version=X.X):
/crowdsourcing/WebServices/get_panel/Epileptic%20encephalopathy/?version=0.1
- Filter the genes by level of confidence:
example 1 (green genes):
/crowdsourcing/WebServices/get_panel/Epileptic%20encephalopathy/?LevelOfConfidence=HighEvidence
example 2 (green genes, red genes):
/crowdsourcing/WebServices/get_panel/Epileptic%20encephalopathy/?LevelOfConfidence=HighEvidence,LowEvidence
- Filter the genes by mode of inheritance (example):
/crowdsourcing/WebServices/get_panel/Epileptic%20encephalopathy/?LevelOfConfidence=HighEvidence,LowEvidence&modesOfInheritance=biallelic
Terms available to use:
-
"MONOALLELIC, autosomal or pseudoautosomal, NOT imprinted": "monoallelic_not_imprinted"
-
"MONOALLELIC, autosomal or pseudoautosomal, maternally imprinted (paternal allele expressed)": "monoallelic_maternally_imprinted"
-
"MONOALLELIC, autosomal or pseudoautosomal, paternally imprinted (maternal allele expressed)": "monoallelic_paternally_imprinted"
-
"MONOALLELIC, autosomal or pseudoautosomal, imprinted status unknown": "monoallelic"
-
"BIALLELIC, autosomal or pseudoautosomal": "biallelic"
-
"BOTH monoallelic and biallelic, autosomal or pseudoautosomal": "monoallelic_and_biallelic"
-
"BOTH monoallelic and biallelic (but BIALLELIC mutations cause a more SEVERE disease form), autosomal or pseudoautosomal": "monoallelic_and_more_severe_biallelic"
-
"X-LINKED: hemizygous mutation in males, biallelic mutations in females": "xlinked_biallelic"
-
"X-LINKED: hemizygous mutation in males, monoallelic mutations in females may cause disease (may be less severe, later onset than males)": "xlinked_monoallelic"
-
"MITOCHONDRIAL": "mitochondrial"
-
"Unknown": "unknown"
11.01.16: Initial gene panels for new nominated rare diseases.
Gene panels for the following diseases have been created on PanelApp, allowing genes to be added by expert review. If you think other existing gene panels should be applied to one of these panels, please contact [email protected].
08.01.16: Changes to some eligibility statements, and new gene panels added.
Initial gene panels for the following diseases have been created on PanelApp, allowing genes to be added by expert review. If you think other existing gene panels should be applied to one of these panels, please contact [email protected].
Ciliopathies
Dermatological disorders
Dysmorphic and congenital abnormality syndromes
Neurology and neurodevelopmental disorders
18.12.15: PanelApp is presented to Dame Una O’Brien, Permanent Secretary at the Department of Health.
15.12.15: A new look for PanelApp is launched to make viewing and reviewing gene panels easier.
07.12.15: We now have 250 reviewers registered!
19.11.15: Gene panels for the following new rare disorders have been added to PanelApp:
13.11.15: The Intellectual disability and Epileptic encephalopathy gene panels were updated to reflect expert reviewer ratings.
03.11.15: Gene panels for the following new rare disorders have been added to PanelApp:
02.11.15: Gene panels are now available to review for the following new rare disorders:
23.10.15: Presentation at the Italian Society of Human Genetics National Congress, Rimini (Ellen McDonagh).
23.10.15: Presentation at the Cardiovascular GeCIP Domain Meeting, London (Augusto Rendon)
22.10.15: We now have 200 experts registered!
21.10.15: Presentation at the GMC/GeCIP Event, London (Ellen McDonagh).
16.10.15: Listen to this week's RARECast (Global Genes) podcast featuring an interview about PanelApp.
15.10.15: PanelApp software version has been updated, making downloads much faster!
01.10.15: PanelApp update
-
Formatted excel spreadsheets of large gene panels (>50 genes) are available for download from the 'Large gene panels' section above, allowing reviewers to add their gene evaluations to the excel spreadsheet in a standardised way and submit their review for upload to PanelApp.
-
An update to the disclaimer was made.
30.09.15: We now have over 100 reviewers registered!
24.09.15: Thank you to all reviews that have been made so far!
View the list of 120 gene panels that still require expert review to find the rare disease in your area of expertise
11.09.15: PanelApp videos released
An introductory video to PanelApp, and another to help Expert Reviewers are released on YouTube.
08.09.15: The PanelApp Handbook PDF is now available from the homepage.
02.09.15: PanelApp Update
- 15 new gene panels are released for each of the new Fast-Track disorders.
- PanelApp is presented at the NHS Expo 2015 in Manchester.
25.08.15: PanelApp presentation at the NHS GMC National Event.
21.08.15: PanelApp is piloted. Version 0 of the gene panels are released for public view and for expert review.
09.04.15: The first line of code for PanelApp is created.
PanelApp is best viewed using Firefox, Chrome or Safari internet browsers. If using Internet Explorer, pop-up information boxes for mode of inheritance, mode of pathogenicity and current diagnostic in the Evaluate Gene tool may not appear. For this information, please refer to the Glossary section under the Contact, Content & Glossary tab on the homepage.
Navigate and Explore PanelApp
PanelApp is an open source tool for gene panel information.
Public users can:
- View gene panels.
- View gene and genomic entity (Short Tandem Repeat/STR and Copy Number Variant/CNV) information.
- Download gene panels.
- View reviewers’ ratings, evaluations and comments and who made these.
- Link to other sources related to the gene such as OMIM (disease-related information), ClinVar (variant-disease related information), and other panels that the gene features on.
- View gene history.
Public users cannot:
- Rate genes and genomic entities (STRs and CNVs) on a gene panel.
- Provide gene evaluations and comments.
- Add genes and genomic entities to panels.
Reviewers can:
- Rate genes and genomic entities (STRs and CNVs) on a gene panel.
- Provide gene evaluations and comments.
- Add genes and genomic entities to panels.
To register as a reviewer, please follow the instructions via the Reviewers tab
Browsing PanelApp as a Public User
Go to the PanelApp home page or to the PanelApp login and click on the ’Browse PanelApp without logging in’ button which will redirect you to the homepage.
Browsing Panels in PanelApp (a Panel-Centric View)
Click on ’Panels’ in the top bar of the page:
A full list of panels is displayed in alphabetical order. The list can be sorted by panel name, percentage of evaluated genes or number of reviewers. The ‘Filter panels’ box allows users to find gene panels of interest, for example; by entering ‘immunodeficiency’, the list will be filtered to display all panels that fall under this term. Click on the link for a gene panel named by disease category (level 4 title).
Comparison of Panels
Two panels can be compared by clicking the ‘Compare two panels’ button and selecting the panels of interest from the drop-down menus. Once selected, click ‘Compare panels’, the genes and their colour rating on each panel will be displayed. For a gene of interest, select ‘Compare’ to see further details regarding the gene on the two panels, including reviews. Please note that this function should not be used for comparing super panels, as it may produce unexpected results.
** Click on the Panel Name to:**
- Read the Genomics England eligibility statement for the disease displayed in the description box.
- View the panel type (Rare Disease 100K, Cancer Germline 100K, External Diagnostic Lab)
- View a list of the reviewers (and their affiliations) who have reviewed genes on this panel.
- View changes to the panel: Click on the 'Panel Activity' button at the bottom of the entity list.
- View the list of genes and genomic entities in the panel, and their current status (e.g. Green, Amber, Red)
- View mode of inheritance (if known), sources the gene was found in, and phenotypes related to the gene.
- View tags attached to genes and entities within a gene panel. These are used by curators to highlight specific information about variants in a gene, for example a ‘nucleotide-repeat-expansion’ tag indicates that nucleotide expansion repeat variations can cause the disorder. See the Glossary section for a full list of entity tags and their definitions.
- From the links at the bottom of the page, you can download the whole panel or a selection of genes and genomic entities.
** Click on a gene symbol to view information about the gene, including: **
HGNC-approved name and links to gene information on Ensembl, OMIM and Gene2Phenotype.
Further information is available on three separate tabs:
- Reviews of the gene.
- Details about the gene including links to ClinVar.
- History lists changes made to the gene information, reviews and status.
** Click on a STR symbol to view information about the STR, including: **
- Chromosome and genomic coordinates for the start and end of the nucleotide repeats in the reference genome for Build 37 and 38.
- The repeated nucleotide sequence
- The maximum number of repeats that are considered non-pathogenic/normal.
- The minimum number of repeats that are considered pathogenic/disease-causing.
- OMIM, Ensembl and Gene2Phenotype information for the corresponding gene.
Further STR information is available on the Reviews, Details and History tabs.
STRs in PanelApp are named as: HGNC-approved gene symbol_Nucleotide repeat (E.g. PPP2R2B_CAG) or the 'STR' tag.
** Click on a CNV symbol to view information about the CNV, including: **
- The CNV identifier (shown in black). This also includes whether the region is a gain or loss; the same region may be present twice on the same panel as a loss and as a gain.
- The CNV full (verbose) name (shown in grey).
- The status of the CNV (Green, Amber, Red) on the panel.
- Chromosome start and end coordinates for GRCh38. For CNVs, only GRCh38 coordinates are provided in PanelApp (unlike for genes and STRs which also display the GRCh37 details).
- The Haploinsufficiency Score (a region loss. This score is provided by ClinGen).
- The Triplosensitivity Score (a region gain. This score is provided by ClinGen).
- Variant type (CNV Loss or CNV Gain)
Further CNV information is available on the Reviews, Details and History tabs.
Note that the ‘Required percent of overlap’ score has a default value of 80%. This score is intended for internal use only.
All CNV symbols are prefixed with 'ISCA' and end with either '-Loss' or '-Gain' (e.g. ISCA-37404-Loss). In the download file, they have the entity type 'region'.
Browsing Genes in PanelApp (a Gene-Centric View)
Click on “Genes and Entities” in the top bar of the PanelApp home page:
A full list of genes and genomic entities will be displayed in alphabetical order. You can select the gene of interest, or enter the symbol into the Filter genes box to narrow down your selection.
Click on a gene symbol to view information about a gene, including:
- HGNC-approved Gene name.
- OMIM gene identifier, and a link to OMIM.
- A link to gene information in Gene2Phenotype.
- A list of all panels on which the gene occurs, and the panel version.
- The status of the gene (Green, Amber, Red) on each panel.
- The Mode of Inheritance of the gene for each disorder (if known).
- Sources in which the gene-phenotype association was found.
- The relevant phenotypes of the gene for each disorder.
From this page, you can:
- Click on the gene symbol to take you to the gene entry within the listed panel.
- Click on the panel name to view the full panel that contains that gene.
Browsing STRs and CNVs in PanelApp from the 'Genes and Entities list.
Short Tandem Repeats (STRs) and Copy Number Variants (CNVs) are searchable from the 'Genes and Entities' tab in the top bar of the PanelApp home page.
If a STR appears on multiple panels, it will appear multiple times in the 'Genes and Entities' list (e.g. ATN1_CAG). Click on the STR to view information about the STR and corresponding gene, and a list of panels the STR and corresponding gene occur on.
All CNV symbols are prefixed with 'ISCA' and end with either '-Loss' or '-Gain' (e.g. ISCA-37404-Loss). Click on a CNV symbol from the 'Genes and Entities' list to view information about the CNV an a list of panels containing the CNV.
Downloading Gene Panels
The download menu is available at the bottom of each panel page. Downloads of panels are in .tsv format, which can be opened using a number of applications, including Microsoft Excel. Users have the option of downloading the (i) whole gene panel, (ii) only Green list, (iii) only Amber list, (iv) Green and Amber list, or (v) only Red list. Users can also download a previous version of the panel. Downloads of the current panel version are also available from the panel page.
The download file contains the following fields:
- Entity Name (gene name, STR name, region name)
- Entity Type (gene, str, region)
- Gene Symbol (HGNC-approved gene symbol)
- Sources (separated by ;)
- Level 4, 3 and 2 titles (E.g. specific rare disease specific, subgroup and group)
- Mode of inheritance (using PanelApp standardised terms)
- Phenotypes (as collected from all sources and reviews)
- OMIM, Orphanet, HPO terms (not currently populated)
- Publications
- Description (not currently populated)
- Flagged (default = FALSE)
- GEL status (reflects the current gene rating; 3 or 4 = Green, 2 = Amber, 1 or 0 = Red)
- User Ratings (represented as the percentage of ratings for Green;Amber;Red).
- Version (the gene panel version)
- Ready
- Mode of pathogenicity
- Ensembl ID (GRCh37)
- Ensembl ID (GRCh38)
- HGNC ID
For STRs only, the following fields will be populated:
- Position Chromosome
- Position GRCh37 start
- Position GRCh37 end
- Position GRCh38 start
- Position GRCh38 end
- STR Repeated Sequence
- STR Normal Repeats
- STR Pathogenic Repeats
For CNVs only, the following fields will be populated:
- Region Haploinsufficiency Score
- Region Triplosensitivity Score
- Region Required Overlap Percentage
- Region Variant Type
- Region Verbose Name
For more detailed guidance about navigating PanelApp please download our PanelApp Handbook V105
PanelApp Reviewers
PanelApp has a crowdsourcing review tool to allow each gene to be reviewed and commented on by experts within the scientific community. Reviewers of PanelApp are worldwide; we currently have more than 500 experts registered from 25 different countries. To become a reviewer, register here
We are asking expert reviewers of the gene panels to help establish a consensus “Green” diagnostic grade list of genes and genomic entities that have a high level of evidence for a role in the relevant rare disease.
Read the PanelApp Reviewers' guide to guide you through the entire review process, from registering to be a reviewer through to leaving a review on a panel.
You can also watch three short videos explaining how to:

Desired reviewer experience
-
Reviewers can have an academic, clinical and/or commercial background.
-
Reviewers can be based anywhere in the world. We currently have registered reviewers from countries including Argentina, Australia, Austria, Bangladesh, Brazil, Canada, France, Germany, Hong Kong, India, Italy, Japan, Korea, Kuwait, Netherlands, New Zealand, Portugal, Qatar, South Korea, Spain, Switzerland, Thailand, Turkey, USA and the UK.
In order to encourage expert review of the gene panels, we would request that reviewers of the gene panels should have:
- Expertise in a disease area, genes or diagnostic genetic testing relevant to the diseases that are part of the NHSE National Genomic Test Directory and Genomic Medicine Service (GMS)
A full range of genomic tests that are commissioned for the NHSE GMS and eligibility criteria of the rare diseases and cancer types included can be found in the National Genomic Test Directory.
What can Reviewers do on PanelApp?
Reviewers can:
- View gene panels.
- View gene information.
- Download gene panels.
- Rate genes in a gene panel.
- Provide gene evaluations and comments.
- View other reviewers’ ratings, evaluations and comments and who made these.
- View a list of their own evaluations.
- Add genes to panels (will be indicated in grey until a Genomics England Curator evaluates the evidence for the gene).
- Link to other sources related to the gene such as OMIM (disease-related information), ClinVar (variant-disease related information).
- View gene history.
Reviewers cannot:
- Delete genes from a panel.
- Delete panels
- Add gene panels
What are we asking of reviewers?
-
Read the eligibility criteria in the National Genomic Test Directory for the clinical indication being reviewed to understand the phenotype criteria established for patient inclusion.
-
Rate the genes and genomic entities on a panel as Green (high evidence, clinically-actionable variants, diagnostically reportable) or Red (low evidence or variants that are not clinically actionable) for the rare disease category, If the evidence is not fully conclusive, please rate as Amber, Please read the criteria for the evidence level required on the Guidelines tab.
-
Provide supporting evidence for your rating, including Publications and Phenotypes.
-
Add missing genes, STRs and CNVs to a gene panel.
Instructions for registering to be a reviewer
Username and password: These are needed to set up and sign in to your reviewer account.
First Name, Last Name and Affiliation: To encourage openness, all gene evaluations and comments from reviewers will be public. Your full name and affiliation will be added to your reviews when you are signed in to your reviewer account. By signing up to be a reviewer, you are agreeing to this condition.
Role, Workplace and Group: This information will not be displayed but will help verify your account.
Email: please use your institute email (e.g. @qmul.ac.uk) to register if possible. This will allow us to process the reviewer request more effectively. If your application is accepted, you will receive confirmation of your reviewer status to this email address.
- After registering, you will be sent an email asking you to validate your email address. Following this a Genomics England curator will review your request and, where appropriate, grant you Reviewer access. In the meantime, you will be signed in with your new username, but will still only have public access and will not be able to make gene evaluations or comments.
Please contact us for any enquires or if you have issues signing in.
How to make a review
Log in
Log in as a reviewer - enter your username and password and click “Log in”.
Read information about PanelApp
You can find out more about PanelApp from the PanelApp homepage, including how gene panels were initially constructed, the role of expert reviewers and gene panel guidelines.
Find gene panels
To find the gene panel(s) assigned to you or relevant to your disease area, click on “Panels” in the top bar of the page. You can see all gene panels listed. The list can be sorted by panel name, number of evaluated genes or number of reviewers. The "Filter panels" box allows users to find gene panels of interest e.g. by entering "diabetes", the list will be filtered to display all panels related to diabetes. For more details on browsing PanelApp, refer to the PanelApp Handbook V105.
Read the eligibility criteria for the rare disease
We request that reviewers read the details regarding the panel in the National Genomic Test Directory and eligibility criteria documentation (where available). The description provides information on the phenotype inclusion and exclusion criteria for the panel.
Review the genes in the gene panel
Click on a gene in the panel and provide a review using the ‘Reviews’ tab (see picture below)
Please leave feedback for each gene on a panel, specifically:
Provide a rating
Provide a gene rating (Green/high evidence/clinically-actionable or diagnostically reportable variants OR Red/low evidence/variants that are not clinically actionable). You may also choose Amber evidence (I don’t know) if it is a borderline case or you are unsure (Guidelines for the evidence required for gene ratings are available via the Guidelines) tab
Provide Free-text justifications in the comments box to support your rating.
If submitting the gene evaluation on behalf of a clinical laboratory, indicate whether variants in the gene are reported as part of current diagnostic practice by checking the 'Clinical diagnostic' box.
Provide mode of inheritance
For each gene, select a mode of inheritance from the drop down menu and submit. Definitions for the terms are provided via the "?" button, and in the Glossary section of the Contact, Content & Glossary tab.
If the mode of inheritance you want to add is not within the drop down menu, or you know of more than one mode of inheritance pattern, different modes of inheritance for specific phenotypes, or other scenarios, please select “other” and provide details in the comments box. Please provide information regarding imprinting, if known.
Provide mode of pathogenicity
We would like to collect exceptions to the rule that loss-of-function variants in this gene can cause the disease. Loss-of-function variants are defined as variants with the sequence ontology terms; transcript ablation, splice acceptor variant, splice donor variant, stop gained, frameshift variant, stop lost, initiator codon variant/start lost.
If loss-of-function variants do not cause the phenotype select “Loss-of-function variants (as defined in pop up message) DO NOT cause this phenotype", and provide details in the comments.
An example of an exception to the rule is the PCSK9 gene, where loss-of-function variants are not relevant for hypercholesterolemia, whereas gain-of-function variant are associated with this phenotype PMID:12730697.
There may be known pathogenic variants in a gene in addition to loss-of-function variants – if a curated set of known-pathogenic variants is available for a gene-phenotype association, please contact us.
Provide additional information to support your review
Phenotypes: Please add phenotypes using standardised OMIM or HPO terms/codes if any additional phenotypes are known to be associated with this gene relevant to the rare disease category (level 4 title). The phenotypes shown in PanelApp are those collected from the sources, with relevance to the disease category (level 4 title).
Publications: Please provide PubMed IDs in the format PMID:12345678;23456789;34567891. Include publications that provide supporting evidence for your given rating e.g. published family pedigree studies, variant reports, functional studies etc supporting the gene-phenotype association. Publications demonstrating a lack of association between the gene and phenotype should also be included.
Comments: Any additional important information, notes of clarification or comments to stimulate debate can be provided in the comments box. Include full references and attributions to sources of information for your review.
Add genes to the Panel
Add any missing genes using the tool found at the bottom of the entity list on the panel (pictured). Provide a rating for the new gene. New genes should be rated Green only if there is significant confidence in reporting in a diagnostic setting (Guidelines for the evidence required for gene ratings are available via the Guidelines tab). Genes that are still in research phase that require further evidence can be added to a panel – select ‘research’ as a source. These should be rated as low evidence (Red).
Note that new genes will be added to the panel as 'Grey' until reviewed by a Genomics England curator.
How to view your evaluations
When you have reviewed a gene, you can see your review under the review tab along with others. A tick will appear against the gene on the left hand side gene list, and on the main panel page genes you have reviewed are also denoted.
When re-visiting PanelApp, click on your username in the top right hand corner of the PanelApp home page to view your user information and a list of your evaluations. Click on the “Go to Gene Evaluations” to make changes or edits to your evaluation, or click on the panel name to view the entire gene panel.
Reviewing STRs and CNVs.
We welcome reviews of STRs and CNVs in PanelApp. Click on the STR or CNV within a panel to leave a review using the 'Reviews' tab. Please provide justifications in the Comments box to support your rating. For STRs, please indicate if interruptions in the repeated sequence are reported as part of standard diagnostic practice in your laboratory/clinic.
You can add STRs and CNVs to the panel using the 'Add a STR to this panel' or 'Add a Region to this panel' button at the bottom of the entity list. However, given that there is a set of minimal information required when adding in an STR or CNV to PanelApp (e.g. genomic coordinates), we suggest that you contact the PanelApp curation team at if you have a STR or CNV you would like to add to the panel.
Considerations when reviewing
- Information for genes on the red list will still be maintained for future reference and may be utilised to prioritise tier 3 variants. As more evidence emerges, may be promoted to the green list.
- Your evaluation and comments will be tagged with your reviewer name and is public.
- The date you made your review will appear, along with the version of the panel you reviewed.
- You can make multiple comments and edit or delete them individually.
- Your review inputs in the review gene tool are saved and so will appear when you log in again.
- Changes to rating, mode of inheritance, mode of pathogenicity and current diagnostic using the gene evaluation tool will overwrite your initial evaluation.
- Publications and phenotypes will be saved in the evaluation tool and can be added to; please note that if you delete what has been saved in these boxes, your original submissions of publications and phenotypes will be overwritten.
**For more detailed guidance about reviewing PanelApp please download our PanelApp Handbook V105
**
For large gene panels or panels for groups of disorders we understand that reviewing using PanelApp may be difficult. We can provide formatted excel sheets of the panels, allowing reviewers to add their reviews to the file for upload to PanelApp. As our panels change over time, please contact us to request the latest version.
This is interim information and has not yet received final approval from Genomics England internal governance processes or NHS England and is therefore potentially subject to change.
PanelApp Gene Panel Guidelines
How to rate the genes
Green (diagnostic-grade) genes included in a Genomics England gene panel for the Rare Disease programme of the 100,000 Genomes Project and NHSE Genomic Medicine Service should fit the criteria A-E outlined below.
These guidelines were developed as a combination of the ClinGen DEFINITIVE evidence for a causal role of the gene in the disease(a), and the Developmental Disorder Genotype-Phenotype (DDG2P) CONFIRMED DD Gene evidence level(b) (please see the original references provided below for full details). These help provide a guideline for expert reviewers when assessing whether a gene should be on the green or the red list of a panel.
A. There are plausible disease-causing mutations(i) within, affecting or encompassing an interpretable functional region(ii) of this gene identified in multiple (3 or more) unrelated cases/families with the phenotype(iii).
OR
B. There are plausible disease-causing mutations(i) within, affecting or encompassing cis-regulatory elements convincingly affecting the expression of a single gene identified in multiple (3 or more) unrelated cases/families with the phenotype(iii).
OR
C. As definitions A or B but in 2 or 3 unrelated cases/families with the phenotype, with the addition of convincing bioinformatic or functional evidence of causation e.g. known inborn error of metabolism with mutation in orthologous gene which is known to have the relevant deficient enzymatic activity in other species; existence of an animal model which recapitulates the human phenotype.
AND
D. Evidence indicates that disease-causing mutations follow a Mendelian pattern of causation appropriate for reporting in a diagnostic setting(iv).
AND
E. No convincing evidence exists or has emerged that contradicts the role of the gene in the specified phenotype.
(i)Plausible disease-causing mutations: Recurrent de novo mutations convincingly affecting gene function. Rare, fully-penetrant mutations - relevant genotype never, or very rarely, seen in controls.
(ii) Interpretable functional region: ORF in protein coding genes miRNA stem or loop.
(iii) Phenotype: the rare disease category, as described in the eligibility statement.
(iv) Intermediate penetrance genes should not be included.
References:
(a) ClinGen Clinical Validity Classifications originally dated July 2014, and updated Oct 2015
A preprint publication is now available: Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the Clinical Genome Resource. Strande et al
(b) The Development Disorder Genotype - Phenotype Database and PMID: 25529582.
NB: On 10th November 2017 the guidelines were updated after clinical approval, changing >3 to 3 or more to reflect curation practices.
Genomics England Gene Panel Principles for Rare Diseases in the 100,000 Genomes Project and NHSE Genomic Medicine Service
- A conservative list of Green genes, STRs and CNVs of known clinical utility and scientific validity is required.
- The Green genes on a panel should be a conservative (diagnostic-grade) set of genes that out of the whole genome should be examined first as variants within these genes are most likely to cause/explain the disease phenotype.
- We acknowledge that the panel will be missing genes that have been reported in association with the disease/phenotype but where the level of proof has not reached that required for them to enter use in a diagnostic setting. Variants that have passed the standard filtering criteria but are not within the gene panel for the relevant disease category will be assigned to a separate tier (Tier 3).
- Genes included on the panel may have been screened in the patient previously; however, with whole genome sequencing, we may find variants of interest not well covered by exome screens or missed by other methods.
- A single gene may appear in multiple gene panels.
- Genes may also be associated with other phenotypes not indicated in the gene panel.
- The gene panels will be updated as we learn from newly published evidence, clinical information, and the 100,000 Genomes Project and NHSE Genomic Medicine Service participant data.
This is interim information and has not yet received final approval from Genomics England internal governance processes or NHS England and is therefore potentially subject to change.
PanelApp API
See our full Swagger API documentation available here
The Swagger PanelApp API can be used to retrieve:
For issues reported with API search functionality please refer to the ‘Known issues in PanelApp functionality and recommended workarounds’ section of the Contact, Content & Glossary page.
Query loading slowly or not completely in your browser? Try adding ?format=json to the end of your query.
PanelApp Frequently Asked Questions
Why gene panels?
The finalised ‘Green’ genes and genomic entities on a Version 1+ gene panel will be used in the tiering of variants to aid clinical interpretation of the genomes sequenced as part of the 100,000 Genomes Project and/or NHSE Genomic Medicine Service. As the virtual gene panels are publicly accessible, they are also available for the scientific and clinical communities.
Why are you asking experts to review the gene panels?
The aim is to utilise expertise and knowledge from the Scientific Community to establish consensus gene panels (a defined green list) for each rare disease category.
Can experts not in the UK or not directly involved in the 100,000 Genomes Project or NHSE Genomic Medicine Service be a reviewer?
Yes! We would like to encourage those with expertise in rare diseases from around the world to register and review genes and gene panels on PanelApp to help gain a consensus view of which genes have enough evidence to be included on a diagnostic virtual gene panel. We currently have more than 500 experts registered from over 25 different countries (including Argentina, Australia, Austria, Bangladesh, Brazil, Canada, France, Germany, Hong Kong, India, Italy, Japan, Korea, Kuwait, Netherlands, New Zealand, Portugal, Qatar, South Korea, Spain, Switzerland, Thailand, Turkey, USA, UK).
As an expert reviewer, how do I rate the genes?
Green Genes included in a Genomics England gene panel for a Rare Disease category for the 100,000 Genomes Project and/or NHSE Genomic Medicine Service should fit the criteria A-E outlined on the Guidelines tab.
Review Scenerios
For the congenital non-syndromic hearing loss (profound/severe) panel
a. A gene is proven to cause syndromic hearing loss but has not to date been proven to manifest with non-syndromic hearing loss. Should it be included in the panel?
Yes, we feel that these genes should be included as a Tier 1 variant in this gene is likely to be relevant to this individual (for example highlighting additional medical features that should be looked for) and should therefore be reported in the genome interpretation.
b. A gene is proven to cause hearing loss, but not congenital profound/severe hearing loss. Should it be included in the panel?
Yes, we feel that these genes should be included as a Tier 1 variant in this gene is likely to be relevant to this individual (even if only partially explaining the phenotype) and should therefore be reported in the genome interpretation.
What will the gene panels and information I provide be used for?
The gene lists and connected information, such as mode of inheritance and pathogenicity, will be used to aid tiering of variants identified in a patient with that disease category, thus will aid genome interpretation and the reporting of clinically-relevant information and may contribute to the diagnosis of the patient. In addition, the gene panels are publicly available and open for research use for the benefit the scientific community.
Who will be able to access the gene panels?
Anyone can view and download the gene panels. In addition, registered reviewers can evaluate genes on the gene panels and provide comments.
Will the gene panels change?
Gene panels may change over time as knowledge accumulates and curators become aware of new evidence. This includes the addition of new genes, Short Tandem Repeats/STRs and Copy Number Variants/CNVs, rating changes to existing genes or genomic entities, and the addition of supporting evidence. The panels may also be merged or split as appropriate, and Super panels may be created made up of two or more child panels. Each change to a panel increases the minor version (E.g. Version 1.0 to Version 1.1). If substantial changes to a gene panel are made, a Genomics England Cuator may increase the gene panel to the next major version (E.g. Version 1.115 to Version 2.0).
Note that Version 1.10 of a gene panel is more recent than Version 1.2, because each minor curation change to the panel increases the minor version incrementally. Think of it instead like Version 1_10 and Version 1_2.
Previous versions of a gene panel can be downloaded using the tool available at the bottom of each gene panel page or by querying API.
The PanelApp ‘Activity’ tab (displayed from the PanelApp home page) displays the last 3000 key changes to all panels. This can be filtered using the filter icon (funnel) to select the panel or date you would like to view. Filter this further by typing in the 'Filter activities' box.
The ‘History’ tab for each gene or genomic entity on a gene panel records changes to gene/entity information. Gene reviews are also added with a date-stamp for tracking.
The 'Panel Activity' tool button at the bottom of the panel description box displays all changes for thiat panel.
Gene panels <Version 1 are viewable and can be reviewed. Once expert reviews have been collected and evaluated internally by Genomics England Curators, Version 1 of the gene panels are released. Version 2 panels will be launched when significant changes have been made to Version 1 panels.
What is recorded when I make my evaluation?
When you are logged in as a reviewer, any information you enter using the gene evaluation or new gene tool will be recorded. Your reviewer name (made up of your first name, last name and affiliation used when you registered to be a reviewer) will be attached to ratings and evaluations. To encourage openness, all gene evaluations and comments you make as a reviewer will be open for anyone to see (the public and other reviewers). The date, time and version of the panel when these actions were carried out is also recorded.
Am I able to change my reviewer evaluations?
As a reviewer, you can log in to PanelApp using your log in details, and view and/or edit your evaluations at any time. To make changes to your evaluations, see Section 1.12.7 of the PanelApp Handbook V105.
We expect PanelApp will still be available to be utilised in between gene panel major version releases in order to collect information. We would also be very appreciative of being contacted regarding important evidence related to gene panels (please contact the curation team at Genomics England:
Who will be assessing the evaluations?
The evaluations will be reviewed by Genomics England Curators using internally established rules and in consultation with Genomics England clinicians.
How will reviews be assessed/conflicting reviews resolved?
A set of rules has been established in order to have a pragmatic review evaluation process. The eligibility criteria from the NHSE National Genomic Test Directory will be used to assess the relevance of a gene on a NHSE Genomic Medicine Service (GMS) panel. Genomics England clinicians will be consulted to resolve conflicting reviews, and expert opinion will be sought from the NHSE GMS, if required. Comments regarding changes to gene ratings are viewable in PanelApp for transparency.
Can I send the link to others so that they can review/download panels?
Please distribute the PanelApp URL or gene panel URLs to those within the Scientific Community; anyone can view and download the gene panels. We also encourage those with expertise to register as reviewers.
Please note; if someone else logs in with your review log in details, any evaluations made will over-ride yours, and comments will not be distinguished as being from more than one user. You are therefore responsible for any changes and comments made under your reviewer name.
When I register as a reviewer, where are my details kept and who has access?
Information added during the registration process are stored internally and only utilised for uses related to PanelApp, it will not be passed to third parties. Your first name, last name, affiliation, workplace and group will be visible to the public. Role is collected in order in order to validate the Reviewer's account.
You can view your account information by clicking on your user name in the top right hand corner of the screen. Please contact the PanelApp team to make any changes to your user information.
Where can I keep up to date with major changes or news regarding PanelApp?
Go to the PanelApp news page. Alternatively, select the ‘Activity’ page from the top menu for the latest key updates to panels. Follow @PanelAppTeam or @GenomicsEngland #PanelApp on Twitter. News items will also be displayed on the Genomics England website – for more information see https://www.genomicsengland.co.uk/about-genomics-england/panelapp/.
Can I submit my own gene panels?
Applications for new panels to be added to the National Genomic Test Directory should be submitted via the NHSE Genomics Unit. For more information, please see the NHS England policy document for Updating the National Genomic Test Directory.
What should I do if I encounter an issue with PanelApp functionality?
We apologise for any inconvenience this may cause. Please first check the ‘Known issues and recommended workarounds’ section on the Contact, Content & Glossary page. If the issue is not listed, we encourage you to contact the PanelApp team to report it.
This is interim information and has not yet received final approval from Genomics England internal governance processes or NHS England and is therefore potentially subject to change.
Contact PanelApp
For enquiries related to PanelApp reviewer accounts, please email: [email protected].
Please note you do NOT need to be registered as a Reviewer to use PanelApp data or download panel content.
For questions and suggestions relating to NHSE Genomic Medicine Service panels, where a review on a panel would not be appropriate, please create a NGIS feedback ticket.
For any other questions or suggestions, please create a request ticket in the Genomics England Service Desk.
Citing PanelApp
To cite PanelApp, please use:
PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels.
Antonio Rueda Martin and Eleanor Williams, Rebecca E. Foulger, Sarah Leigh, Louise C. Daugherty, Olivia Niblock, Ivone U. S. Leong, Katherine R. Smith, Oleg Gerasimenko, Eik Haraldsdottir, Ellen Thomas, Richard H. Scott, Emma Baple, Arianna Tucci, Helen Brittain, Anna de Burca, Kristina Ibañez, Dalia Kasperaviciute, Damian Smedley, Mark Caulfield, Augusto Rendon & Ellen M. McDonagh. Nat Genet (2019) doi:10.1038/s41588-019-0528-2. PubMed
Where applicable, please provide the name and version of the gene panel(s).
Terms of Use
Please find the attached Genomics England PanelApp Terms of Use, Dec 2019
Known issues in PanelApp functionality and recommended workarounds
The following issues have been reported by other PanelApp users. We are working to resolve these issues alongside other priorities.
PanelApp API
- Genes endpoint (/api/v1/genes/) - This endpoint returns inconsistent results which vary in order with each query. We advise using the entities endpoint (/api/v1/entities/) instead and filtering for genes.
- Panel versions endpoint (/api/v1/panels/{id}/versions) - This endpoint currently returns a status 500 error. To retrieve a specific panel version, include the version number in the query. For example:
GET /api/v1/panels/{id}/?version=1.3 will return version 1.3 of the panel.
- Panels endpoint (/api/v1/panels/) - Searching by panel name (e.g. intellectual disability) may return unexpected results in some cases due to the fuzzy matching algorithm used. If you wish to ensure accuracy, please use the panel ID (e.g. 285) instead of the panel name. This is a feature of this search and not a bug and we will not be changing this behaviour.
PanelApp User Interface
- Compare two panels function - The comparison tool on the Panels page does not always return the highest ranked instance of a gene if the comparison includes a super panel. We do not recommend using this function for super panel comparisons.
- Gene search box behaviour - On the Genes and Entities page, pressing Enter after typing or pasting a gene name in the 'Filter genes and genomic entities' search box no longer initiates a search. Please click the gene name displayed below the search box to proceed.
- STR-linked gene panel count - The number of panels associated with a gene linked to an STR is incorrectly shown as 0 on STR pages (e.g. ATXN1_CAG). To view the correct list, click the ‘0 panels’ text.
- Download Latest Signed-Off Version - The 'Download Latest Signed-Off Version' button at the top and bottom of some panel pages may result in a 500 error (e.g. Hereditary neuropathy or pain disorder). As a workaround, you can:
- Visit the NHS GMS Panels site to view the latest signed-off version of the panel.
- Obtain panel content via the API panel versions endpoint (e.g. api/v1/panels/846/?version=7.0)
Acknowledgements
Sources of information
Please refer to the original sources for more information, including disclaimers and usage terms.
The 4 sources used in the initial establishment of gene lists are:
Please see the sources for more information, including disclaimers and use of information. Information from these sources (manually collected from January 2015) includes:
- Genes.
- Phenotypes.
- Mode of inheritance (predominantly from the Illumina TruGenome Predisposition Screen, but also captured from UKGTN information and Emory where available).
-
Transcripts (predominantly from the Illumina TruGenome Predisposition Screen).
-
Gene mapping information is sourced from Ensembl version 94. Reference: Zerbino DR et al. Ensembl 2018. Nucleic Acids Res. 46(D1):D754-D761. (2018).
Other databases used as resources used for some phenotype, gene or mode of inheritance information and to aid the curation process:
-
Online Mendelian Inheritance in Man - OMIM. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD).
-
Gene2Phenotype. References: Wright CF et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 385(9975):1305-14. (2015), The Development Disorder Genotype - Phenotype Database.
-
Orphanet
-
The NCBI Genetics Home Reference
-
The HGNC website is used to check gene names in order to establish the correct HGNC-approved symbol. Gene terms used within PanelApp are HGNC approved names. Reference: Braschi B. et al. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res. 47(D1):D786-D792. (2019).
-
ClinGen Gene Validity Curations. Reference: Strande NT et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 100(6):895-906. (2017).
Other sources of gene lists
-
Diagnostic-grade genes in the The Victorian Clinical Genetics Services were used to revise and re-evaluate PanelApp gene panels. Thank you to Dr Zornitza Stark for providing these lists and reviews.
-
The NIHR BioResource- Rare Diseases (Bridge) Study was used to revise and re-evaluate PanelApp gene panels. PanelApp displays the Source: 'BRIDGE study Tier 1 gene' for these genes.
-
ClinGen Gene Validity Curations. Reference: Strande NT et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 100(6):895-906. (2017).
-
The Simons Foundation Autism Research Initiative (SFARI) Gene database.
CNV and STR information
-
Regions in PanelApp were mapped from the curated ClinGen Dosage Sensitivity Map curated regions, available from here. The Clinical Genome Resource (ClinGen) consortium is curating genes and regions of the genome to assess whether there is evidence to support that these genes/regions are dosage sensitive and should be targeted on a cytogenomic array. Missing genomic coordinates were provided by Dalia Kasperaviciute (Genomics England).
-
STR information was provided by Arianna Tucci, Kristina Ibanez-Garikano and Katherine Smith (Genomics England).
Link outs from PanelApp
-
Links to ClinVar from gene pages are provided. Reference: Landrum M.J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44(D1):D862-8. (2016).
-
Links to Gene2Phenotype from gene pages are provided. References: Wright CF et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 385(9975):1305-14. (2015), The Development Disorder Genotype - Phenotype Database.
-
Links to OMIM from gene pages are provided.
-
Links are provided to Ensembl Build 37 and Build 38 from gene pages. Reference: Zerbino DR et al. Ensembl 2018. Nucleic Acids Res. 46(D1):D754-D761. (2018).
Other source types:
-
Expert list or Expert: this source in PanelApp indicates that the gene was submitted on a list of genes from an expert in the disease area.
-
Expert Review: this source in PanelApp indicates that genes were added by an expert during review of the gene panels.
-
Literature: this source in PanelApp indicates that the gene was sourced from a peer-reviewed published article. Links to PubMed via PubMed IDs (PMIDs) as a reference to publications are provided where possible.
-
Eligibility statement prior genetic testing: this source in PanelApp indicates that prior genetic testing is required/advised on the Genomics England Eligibility Statement for that disorder. The eligibility statements are available here.
PanelApp contributors
Thank you to all participants and partners of the 100,000 Genomes Project
** Thank you to all reviewers and those who have contributed feedback to help the development of PanelApp. Please see individual panels for the name and affiliation of contributing reviewers.**
** Thank you to all experts who contributed gene lists during the development of rare disease eligibility statements and data models, or by submitting their own gene panels. These have been added to the gene panels on PanelApp where possible.**
** Thank you to the NHSE Genomic Medicine Service disease specialist groups for providing gene lists and reviews.**
PanelApp creator:
Antonio Rueda-Martin
PanelApp Product Owner:
Todd Donnelly
PanelApp Developers: Alasdair Colley, Antonio Rueda-Martin, Oleg Gerasimenko.
Current PanelApp Curators:
Eleanor Williams, Achchuthan Shanmugasundram, Arina Puzriakova, Ida Ertmanska.
Past PanelApp Curators: Ellen McDonagh, Sarah Leigh, Catherine Snow, Ivone Leong, Rebecca Foulger, Louise Daugherty, Mafalda Gomes.
Other contributers to PanelApp curation, development, content, documentation and outreach:
Eik Haraldsdottir, Ellen Thomas, Caroline Wright, Emma Baple, Damian Smedley, Chris Boustred, Kirsty McCaffrey, Alice Gardham, Richard Scott, Chris Campbell, Rachel Jones, Olivia Niblock, Anna de Burca, Helen Brittain, Arianna Tucci, Augusto Rendon, Katherine Smith, Clare Turnbull, Jo Whittaker, Mina Ryten, Tom Fowler, Mark Caulfield, Andrew Devereau, Alona Sosinksy, Maria Asthanasopoulou, Kristina Ibanez-Garikano, Dalia Kasperaviciute, Corey Johnson, James Carroll, Lisa Carr, Lisa Dinh, Verity Fryer, Paul Hayes, James Carroll, Matthew Parker, Lara Hawkes, Genomics England Platform Team & Service Desk, Members of the E&I GeCIP Domain (Sian Ellard, Steve Abbs, Dominic McCullun, Helen Firth, William Newman), Carrie Mowatt, Samuel Barnett, Thomas Erker, Deepti Sood, Cassandra Smith, Nour Elkhateeb, Susan Walker, Maricica Zabrautanu, Jane Deller, Rachael Mein, Anna Lisa Taylor Tavares, Zerin Hyder, Meriel McEntagart.
PanelApp Glossary
Retired panels
Some gene panels that were previously used for genome analysis and interpretation have now been retired. These panels can no longer be found through the Panels page in the user interface or through the panels query of the API (/api/v1/panels/).
However, users can still access older versions if needed:
- Via the API: by specifying the panel ID and version number (e.g. /api/v1/panels/473/?version=3.0)
- Via the user interface: by entering the panel ID directly in the URL (e.g. R147 Growth failure in early childhood)
Level 4 title, Level 3 title, Level 2 title
These terms refer to the rare disorder categories and titles e.g. Arrhythmogenic Right Ventricular Cardiomyopathy (level 4 title) is a disease under the cardiomyopathies sub-category (level 3 title), within cardiovascular disorders (level 2 title).
Panel Type
This indicates what the gene panel is used for:
-
Cancer Germline 100K: a panel used for the interpretation pipeline for cancer genomes from the 100,000 Genomes Project.
-
Rare Disease 100K: a panel used for the interpretation pipeline for rare disease genomes from the 100,000 Genomes Project.
-
GMS Cancer Germline Virtual: a panel developed for analysis of cancer susceptibility findings in the Genomic Medicine Service for WGS clinical indications in the National Genomic Test Directory for cancer.
-
GMS Rare Disease: a panel developed for the NHS Genomic Medicine Service in England - may be delivered by whole exome sequencing, large or small 'wet lab' panel.
-
GMS Rare Disease Virtual: a panel developed for the NHS Genomic Medicine Service in England that will be used as a virtual panel for whole genome sequencing indications.
-
GMS Signed-off: this gene panel has been signed-off by NHSE governance processes for the Genomic Medicine Service.
-
Super Panel: a panel that is composed of multiple component panels. When updates to component panels are made, the super panel is automatically updated.
-
Component Of Super Panel: a panel that has been added to a super panel. Changes to this panel will automatically be updates on the super panel.
-
Actionable: a panel containing actionable-type information linked to genes, such as clinical trial information.
-
Research: a panel used for research purposes.
Relevant disorders
Test codes (e.g. R codes) from the NHS England National Genomic Test Directory and previous panel and/ or clinical indication names that were used in earlier versions of the test directory are added as relevant disorders for the NHSE Genomic Medicine Service panels.
Panels that originated in the 100,000 Genomes Project may also list diseases relevant to the panel in the Relevant disorders field.
Sometimes gene panels are merged to form a new panel encompassing a broader disease spectrum - the original individual panels will be retired, and the panel names added as relevant disorders to the new merged panel.
Eligibility statement
The eligibility statements for gene panels used in tests that are part of the NHS England Genomic Medicine Service can be found in the eligibility criteria document from the National Genomic Test Directory website.
The eligibility statements for each of the rare diseases that were tested as part of the 100,000 Genomes Project can be found on the Genomics England Website.
Entity
A gene, Short Tandem Repeat (STR) or region (CNV) on a gene panel.
STR
Short Tandem Repeat. These have 'Entity type:str' in PanelApp. For further information on what STR information is captured, refer to Section 1.9.7 in the PanelApp Handbook V105.
CNV
Copy Number Variant. These have 'Entity type:region' in PanelApp. For further information on what CNV information is captured, refer to Section 1.9.8 in the PanelApp Handbook V105.
Gene Symbol
The HGNC-approved symbol for the gene from Ensembl release 90.
Gene Name
The HGNC-approved name for the gene from Ensembl release 90.
Gel Status
This provides the overall rating of the gene, based on either the initial sources or curated evidence.
For panels that are version 0:
- 3 or 4 = Green = from 3 or 4 of the original sources used to establish an initial gene list, as listed above.
- 2 = Amber = from 2 of the original sources.
- 1 = Red = from 1 of the original sources.
- 0 = Red = a gene from a submitted list or other source.
For panels that are version 1+ this corresponds to:
- 3 or 4 = Green = highest level of confidence/evidence for the gene-disease. These genes are used for genome interpretation.
- 2 = Amber = moderate level of evidence
- 0 or 1 = Red = lowest level of evidence
Mode of Inheritance
Standardised terms were used to represent the gene-disease mode of inheritance, and were mapped to commonly used terms from the different sources. Below each of the terms is described, along with the equivalent commonly-used terms.
MONOALLELIC, autosomal or pseudoautosomal, not imprinted: A variant on one allele of this gene can cause the disease, and imprinting has not been implicated.
MONOALLELIC, autosomal or pseudoautosomal, maternally imprinted (paternal allele expressed): A variant on the paternally-inherited allele of this gene can cause the disease, if the alternate allele is imprinted (function muted).
MONOALLELIC, autosomal or pseudoautosomal, paternally imprinted (maternal allele expressed): A variant on the maternally-inherited allele of this gene can cause the disease, if the alternate allele is imprinted (function muted).
MONOALLELIC, autosomal or pseudoautosomal, imprinted status unknown: A variant on one allele of this gene can cause the disease. This is the default used for autosomal dominant mode of inheritance where no knowledge of the imprinting status of the gene required to cause the disease is known. Mapped to the following commonly used terms from different sources: autosomal dominant, dominant, AD, DOMINANT.
BIALLELIC, autosomal or pseudoautosomal: A variant on both alleles of this gene is required to cause the disease. Mapped to the following commonly used terms from different sources: autosomal recessive, recessive, AR, RECESSIVE.
BOTH monoallelic and biallelic, autosomal or pseudoautosomal: The disease can be caused by a variant on one or both alleles of this gene. Mapped to the following commonly used terms from different sources: autosomal recessive or autosomal dominant, recessive or dominant, AR/AD, AD/AR, DOMINANT/RECESSIVE, RECESSIVE/DOMINANT.
BOTH monoallelic and biallelic, autosomal or pseudoautosomal (but BIALLELIC mutations cause a more SEVERE disease form), autosomal or pseudoautosomal: A variant on one allele of this gene can cause the disease, however a variant on both alleles of this gene can result in a more severe form of the disease/phenotype.
X-LINKED: hemizygous mutation in males, biallelic mutations in females: A variant in this gene can cause the disease in males as they have one X-chromosome allele, whereas a variant on both X-chromosome alleles is required to cause the disease in females. Mapped to the following commonly used term from different sources: X-linked recessive, XLR, hemizygous.
X linked: hemizygous mutation in males, monoallelic mutations in females may cause disease (may be less severe, later onset than males): A variant in this gene can cause the disease in males as they have one X-chromosome allele. A variant on one allele of this gene may also cause the disease in females, though the disease/phenotype may be less severe and may have a later-onset than is seen in males. X-linked inactivation and mosaicism in different tissues complicate whether a female presents with the disease, and can change over their lifetime. This term is the default setting used for X-linked genes, where it is not known definitely whether females require a variant on each allele of this gene in order to be affected. Mapped to the following commonly used terms from different sources: X-linked dominant, XLD, x-linked, X-LINKED, X-linked.
MITOCHONDRIAL: The gene is in the mitochondrial genome and variants within this can cause this disease, maternally inherited. Mapped to the following commonly used term from different sources: Mitochondrial.
Unknown: Mapped to the following commonly used terms from different sources: Unknown, NA, information not provided.
Other - please specify in evaluation comments: For example, if the mode of inheritance is digenic, please indicate this in the comments and which other gene is involved.
Phenotypes
Phenotypes collected from the sources are provided where possible. Where multiple phenotypes for the gene were listed, it may be that only the top phenotype was captured or only the relevant phenotype for the gene panel was collected. Phenotypes may also be sourced from PanelApp reviewers, OMIM or Gene2Phenotype during curation of gene panels.
OMIM
A link to the gene page on OMIM is provided to give reviewers quick access to gene-disease information.
OMIM SYMBOLS
Brackets, "[ ]" indicate "nondiseases," mainly genetic variations that lead to apparently abnormal laboratory test values (e.g., dysalbuminemic euthyroidal hyperthyroxinemia).
Braces, "{ }" indicate mutations that contribute to susceptibility to multifactorial disorders (e.g., diabetes, asthma) or to susceptibility to infection (e.g., malaria).
A question mark, "?" before the phenotype name indicates that the relationship between the phenotype and gene is provisional. More details about this relationship are provided in the comment field of the map and in the gene and phenotype OMIM entries.
The number in parentheses after the name of each disorder indicates the following: (1) the disorder was positioned by mapping of the wildtype gene; (2) the disease phenotype itself was mapped; (3) the molecular basis of the disorder is known; (4) the disorder is a chromosome deletion or duplication syndrome. Move the cursor over the number to display this information.
ClinVar Variants
A link to the gene page on Clinvar is provided for quick access to information regarding variants within the gene that are associated with different conditions.
Penetrance
This is set as a default to "complete". Please provide information regarding the penetrance in the Comments box in the Evaluate Gene tool.
Publications
Publications that provide evidence linking this gene to this disorder.
Mode of Pathogenicity
For each gene in a gene panel in PanelApp, it is assumed that loss-of-function variants in this gene can cause the disease/phenotype unless an exception to this rule is known. In the PanelApp, we would like to collect information regarding exceptions to this rule. An example of an exception is the PCSK9 gene, where loss-of-function variants are not relevant for a hypercholesterolemic phenotype as they are associated with increased LDL-cholesterol uptake via LDLR PMID: 25911073.
In the PanelApp, we classify loss-of-function variants as those with the following Sequence Ontology (SO) terms:
transcript ablation, splice acceptor variant, splice donor variant, stop gained, frameshift variant, stop lost, initiator codon variant/start lost.
SO Terms and descriptions
Sourced from Ensembl:
transcript ablation: A feature ablation whereby the deleted region includes a transcript feature (SO:0001893)
splice acceptor variant: A splice variant that changes the 2 base region at the 3' end of an intron (SO:0001574)
splice donor variant: A splice variant that changes the 2 base region at the 5' end of an intron (SO:0001575)
stop gained: A sequence variant whereby at least one base of a codon is changed, resulting in a premature stop codon, leading to a shortened transcript (SO:0001587)
frameshift variant: A sequence variant which causes a disruption of the translational reading frame, because the number of nucleotides inserted or deleted is not a multiple of three (SO:0001589)
stop lost: A sequence variant where at least one base of the terminator codon (stop) is changed, resulting in an elongated transcript (SO:0001578)
initiator codon variant: A codon variant that changes at least one base of the first codon of a transcript/start lost: a codon variant that changes at least one base of the canonical start codon (SO:0001582)
This gene appears in other panels
A list of other gene panels within PanelApp that contain this gene.
Rating Summary
A summary of the ratings by different reviewers for the level of evidence for this gene to be included on this panel.
Gene History/Genomic Entity History
A record of when the gene or genomic entity (a STR or CNV) was added to the panel in PanelApp and additional key changes to gene or entity information.
Current diagnostic
This is a checkbox for reviewers available in the Evaluate Gene tool to indicate whether or not they report variants in the gene as part of current diagnostic practice. If you are submitting an evaluation on behalf of a clinical laboratory and you report variants within the gene as part of your current diagnostic practice, please check the box.
Tags
Are attached to a gene or genomic entity within a gene panel by a curator. Tags highlight useful information about a gene or gene variants that may affect gene ratings or be useful for future curation. Tags are specific to a gene within a given panel. A list of tags and descriptions is available here.






























 A review of the year up to July 2018
A review of the year up to July 2018






























